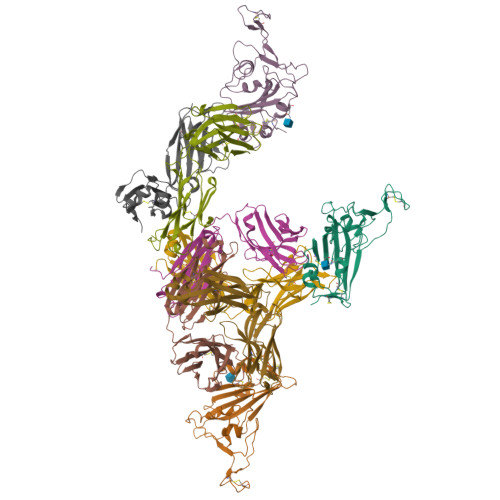
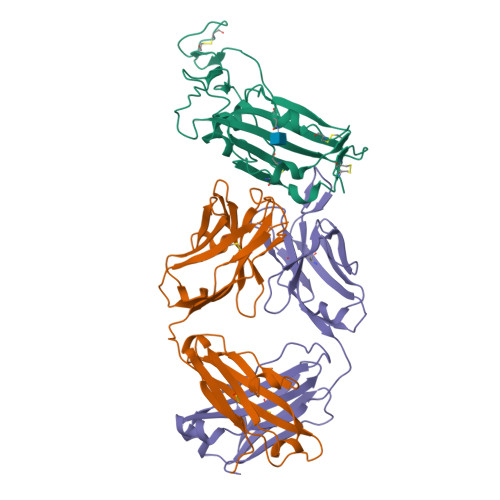
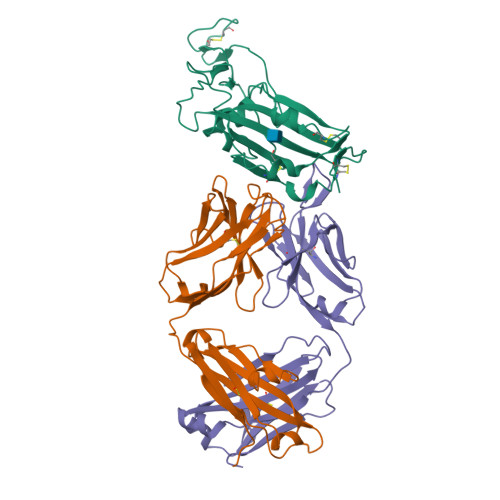
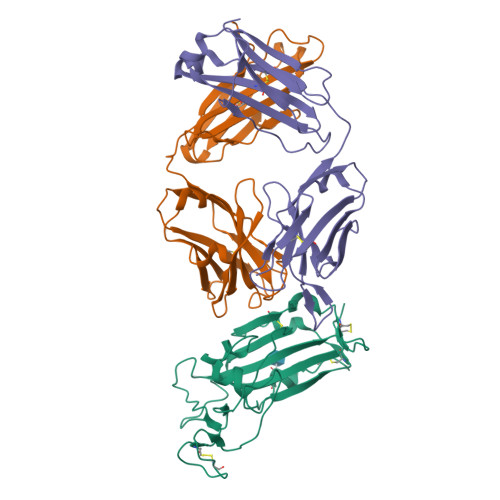
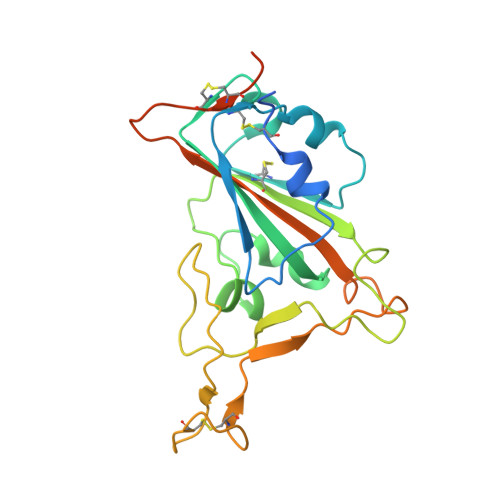
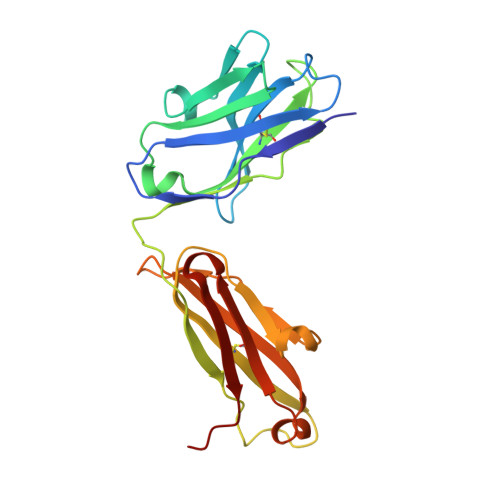
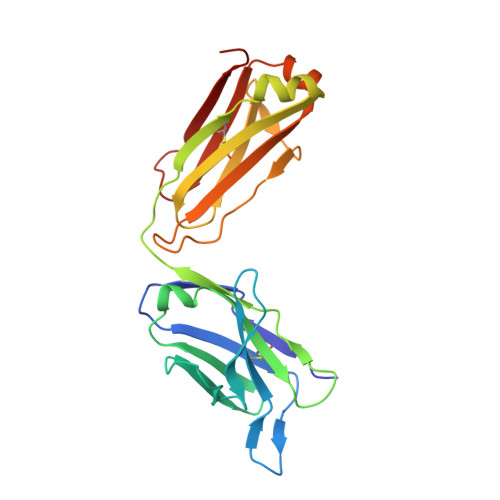
A New Crystal Form of the SARS-CoV-2 Receptor Binding Domain: CR3022 Complex-An Ideal Target for In-Crystal Fragment Screening of the ACE2 Binding Site Surface.
Nichols, C., Ng, J., Keshu, A., Fraternali, F., De Nicola, G.F.(2020) Front Pharmacol 11: 615211-615211
- PubMed: 33381049
- DOI: https://doi.org/10.3389/fphar.2020.615211
- Primary Citation of Related Structures:
6ZLR - PubMed Abstract:
In-crystal fragment screening is a powerful tool to chemically probe the surfaces used by proteins to interact, and identify the chemical space worth exploring to design protein-protein inhibitors. A crucial prerequisite is the identification of a crystal form where the target area is exposed and accessible to be probed by fragments. Here we report a crystal form of the SARS-CoV-2 Receptor Binding Domain in complex with the CR3022 antibody where the ACE2 binding site on the Receptor Binding Domain is exposed and accessible. This crystal form of the complex is a valuable tool to develop antiviral molecules that could act by blocking the virus entry in cells.
Organizational Affiliation:
British Heart Foundation Centre of Excellence, Department of Cardiology, Rayne Institute, St Thomas' Hospital, King's College London, London, United Kingdom.