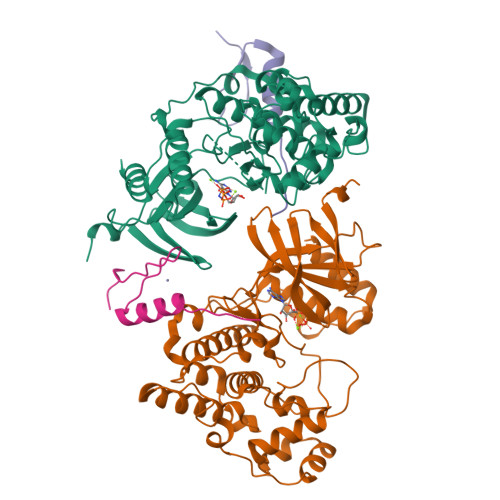
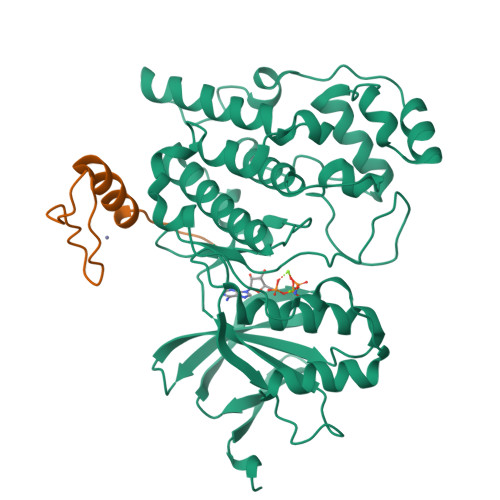
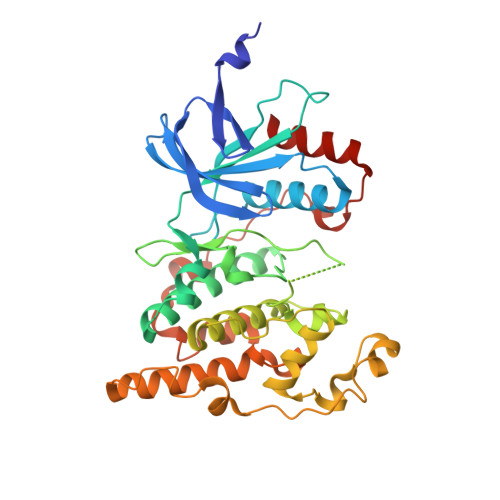
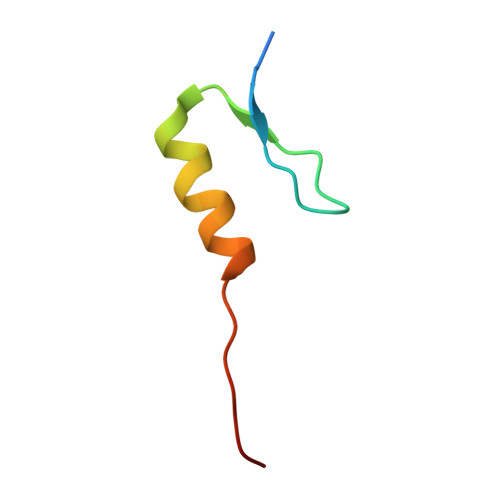
Co-regulation of the transcription controlling ATF2 phosphoswitch by JNK and p38.
Kirsch, K., Zeke, A., Toke, O., Sok, P., Sethi, A., Sebo, A., Kumar, G.S., Egri, P., Poti, A.L., Gooley, P., Peti, W., Bento, I., Alexa, A., Remenyi, A.(2020) Nat Commun 11: 5769-5769
- PubMed: 33188182
- DOI: https://doi.org/10.1038/s41467-020-19582-3
- Primary Citation of Related Structures:
6ZQS, 6ZR5 - PubMed Abstract:
Transcription factor phosphorylation at specific sites often activates gene expression, but how environmental cues quantitatively control transcription is not well-understood. Activating protein 1 transcription factors are phosphorylated by mitogen-activated protein kinases (MAPK) in their transactivation domains (TAD) at so-called phosphoswitches, which are a hallmark in response to growth factors, cytokines or stress. We show that the ATF2 TAD is controlled by functionally distinct signaling pathways (JNK and p38) through structurally different MAPK binding sites. Moreover, JNK mediated phosphorylation at an evolutionarily more recent site diminishes p38 binding and made the phosphoswitch differently sensitive to JNK and p38 in vertebrates. Structures of MAPK-TAD complexes and mechanistic modeling of ATF2 TAD phosphorylation in cells suggest that kinase binding motifs and phosphorylation sites line up to maximize MAPK based co-regulation. This study shows how the activity of an ancient transcription controlling phosphoswitch became dependent on the relative flux of upstream signals.
Organizational Affiliation:
Biomolecular Interactions Research Group, Institute of Organic Chemistry, Research Center for Natural Sciences, H-1117, Budapest, Hungary.