Inhibition of Urease, a Ni-Enzyme: The Reactivity of a Key Thiol With Mono- and Di-Substituted Catechols Elucidated by Kinetic, Structural, and Theoretical Studies.
Mazzei, L., Contaldo, U., Musiani, F., Cianci, M., Bagnolini, G., Roberti, M., Ciurli, S.(2021) Angew Chem Int Ed Engl 60: 6029-6035
- PubMed: 33245574
- DOI: https://doi.org/10.1002/anie.202014706
- PubMed Abstract:
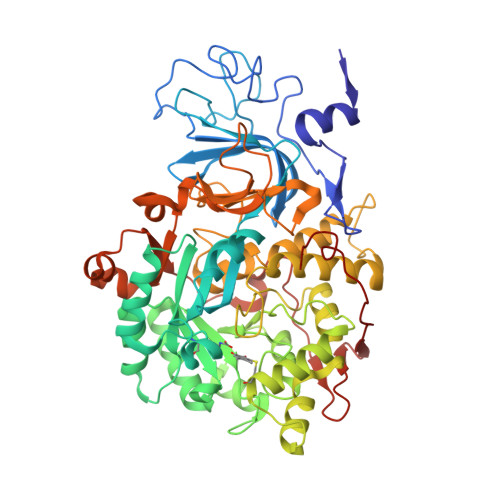
The inhibition of urease from Sporosarcina pasteurii (SPU) and Canavalia ensiformis (jack bean, JBU) by a class of six aromatic poly-hydroxylated molecules, namely mono- and dimethyl-substituted catechols, was investigated on the basis of the inhibitory efficiency of the catechol scaffold. The aim was to probe the key step of a mechanism proposed for the inhibition of SPU by catechol, namely the sulfanyl radical attack on the aromatic ring, as well as to obtain critical information on the effect of substituents of the catechol aromatic ring on the inhibition efficacy of its derivatives. The crystal structures of all six SPU-inhibitors complexes, determined at high resolution, as well as kinetic data obtained on JBU and theoretical studies of the reaction mechanism using quantum mechanical calculations, revealed the occurrence of an irreversible inactivation of urease by means of a radical-based autocatalytic multistep mechanism, and indicate that, among all tested catechols, the mono-substituted 3-methyl-catechol is the most efficient inhibitor for urease.
Organizational Affiliation:
Department of Pharmacy and Biotechnology (FaBiT), University of Bologna, Via Belmeloro 6, 40126, Bologna, Italy.