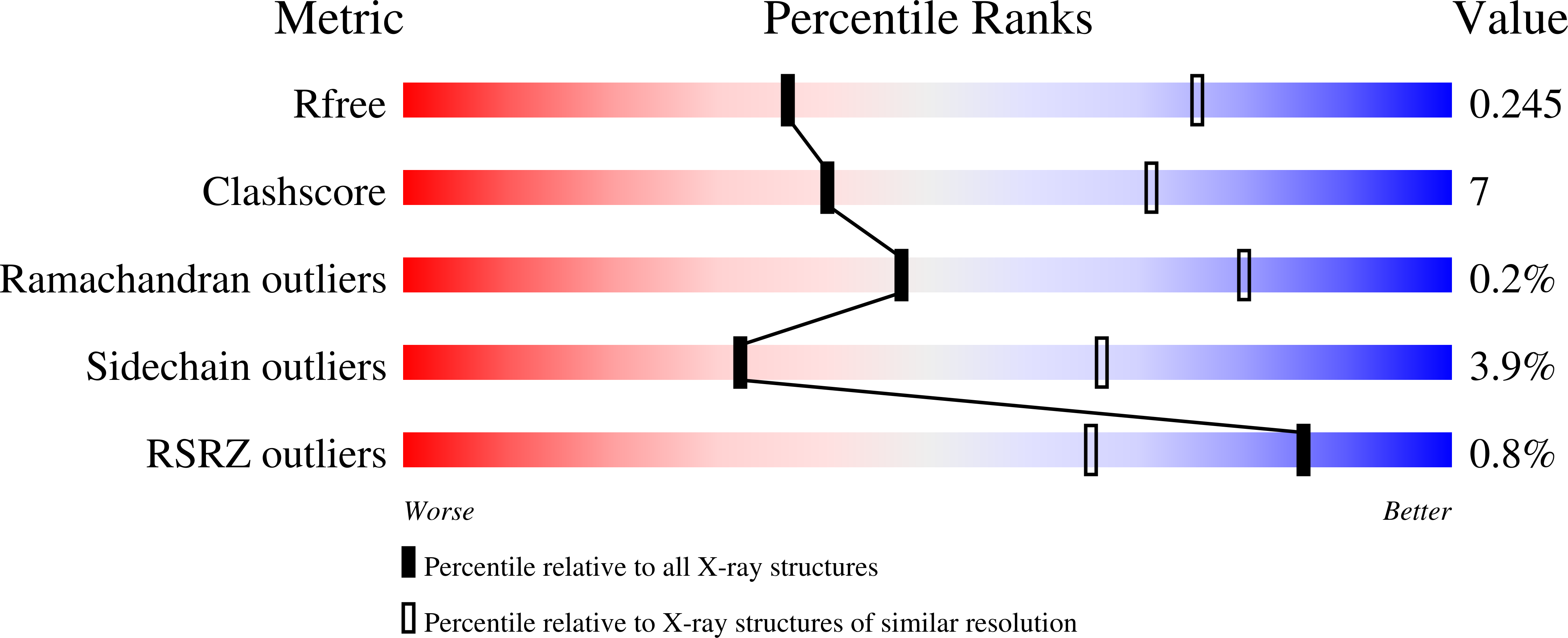
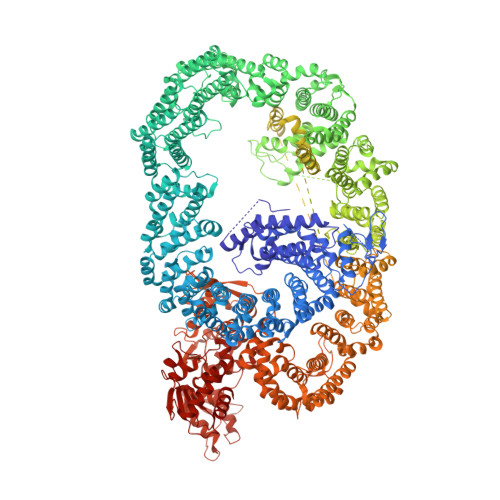
HUWE1 employs a giant substrate-binding ring to feed and regulate its HECT E3 domain.
Grabarczyk, D.B., Petrova, O.A., Deszcz, L., Kurzbauer, R., Murphy, P., Ahel, J., Vogel, A., Gogova, R., Faas, V., Kordic, D., Schleiffer, A., Meinhart, A., Imre, R., Lehner, A., Neuhold, J., Bader, G., Stolt-Bergner, P., Bottcher, J., Wolkerstorfer, B., Fischer, G., Grishkovskaya, I., Haselbach, D., Kessler, D., Clausen, T.(2021) Nat Chem Biol 17: 1084-1092
- PubMed: 34294896
- DOI: https://doi.org/10.1038/s41589-021-00831-5
- Primary Citation of Related Structures:
7BII - PubMed Abstract:
HUWE1 is a universal quality-control E3 ligase that marks diverse client proteins for proteasomal degradation. Although the giant HECT enzyme is an essential component of the ubiquitin-proteasome system closely linked with severe human diseases, its molecular mechanism is little understood. Here, we present the crystal structure of Nematocida HUWE1, revealing how a single E3 enzyme has specificity for a multitude of unrelated substrates. The protein adopts a remarkable snake-like structure, where the C-terminal HECT domain heads an extended alpha-solenoid body that coils in on itself and houses various protein-protein interaction modules. Our integrative structural analysis shows that this ring structure is highly dynamic, enabling the flexible HECT domain to reach protein targets presented by the various acceptor sites. Together, our data demonstrate how HUWE1 is regulated by its unique structure, adapting a promiscuous E3 ligase to selectively target unassembled orphan proteins.
Organizational Affiliation:
Research Institute of Molecular Pathology, Vienna BioCenter, Vienna, Austria. daniel.grabarczyk@imp.ac.at.