A Novel Therapeutic Anti-ErbB3, ISU104 Exhibits Potent Antitumorigenic Activity by Inhibiting Ligand Binding and ErbB3 Heterodimerization.
Hong, M., Yoo, Y., Kim, M., Kim, J.Y., Cha, J.S., Choi, M.K., Kim, U., Kim, K., Sohn, Y., Bae, D., Cho, H.S., Hong, S.B.(2021) Mol Cancer Ther 20: 1142-1152
- PubMed: 33782100
- DOI: https://doi.org/10.1158/1535-7163.MCT-20-0907
- Primary Citation of Related Structures:
7D85 - PubMed Abstract:
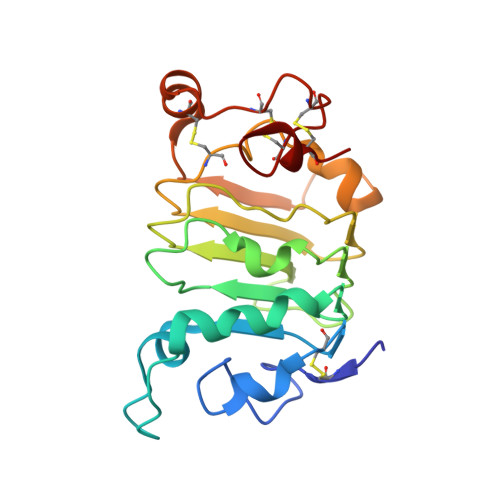
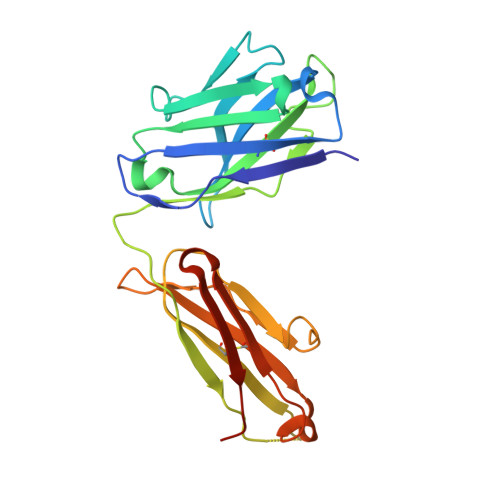
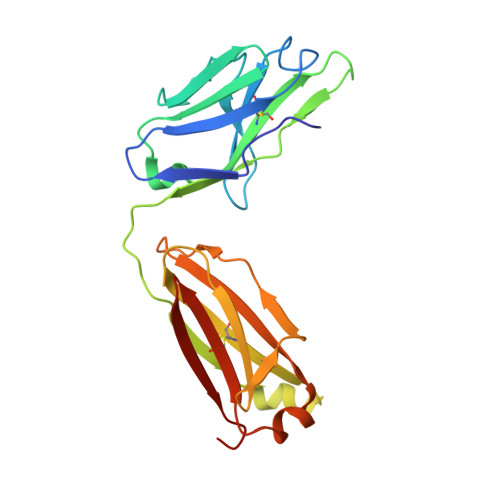
ErbB3, a member of the ErbB receptor family, is a potent mediator in the development and progression of cancer, and its activation plays pivotal roles in acquired resistance against anti-EGFR therapies and other standard-of-care therapies. Upon ligand (NRG1) binding, ErbB3 forms heterodimers with other ErbB proteins (i.e., EGFR and ErbB2), which allows activation of downstream PI3K/Akt signaling. In this study, we developed a fully human anti-ErbB3 antibody, named ISU104, as an anticancer agent. ISU104 binds potently and specifically to the domain 3 of ErbB3. The complex structure of ErbB3-domain 3::ISU104-Fab revealed that ISU104 binds to the NRG1 binding region of domain 3. The elucidated structure suggested that the binding of ISU104 to ErbB3 would hinder not only ligand binding but also the structural changes required for heterodimerization. Biochemical studies confirmed these predictions. ISU104 inhibited ligand binding, ligand-dependent heterodimerization and phosphorylation, and induced the internalization of ErbB3. As a result, downstream Akt phosphorylation and cell proliferation were inhibited. The anticancer efficacy of ISU104 was demonstrated in xenograft models of various cancers. In summary, a highly potent ErbB3 targeting antibody, ISU104, is suitable for clinical development.
Organizational Affiliation:
Research Institute, ISU ABXIS Co., Ltd., Sungnam-si, Republic of Korea (South).