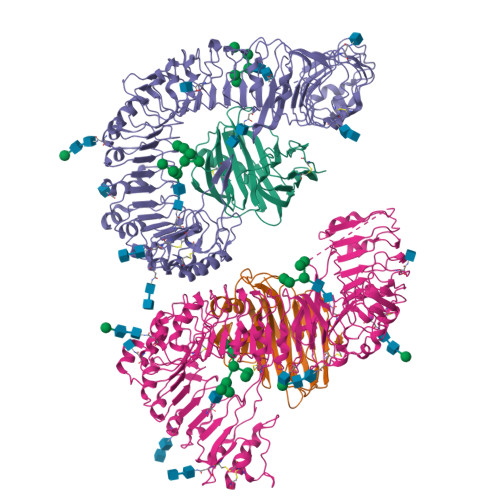
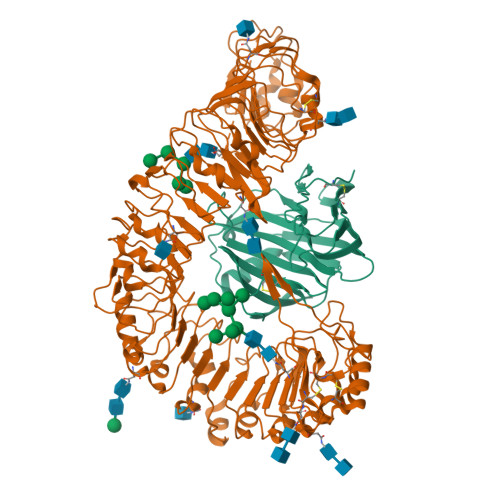
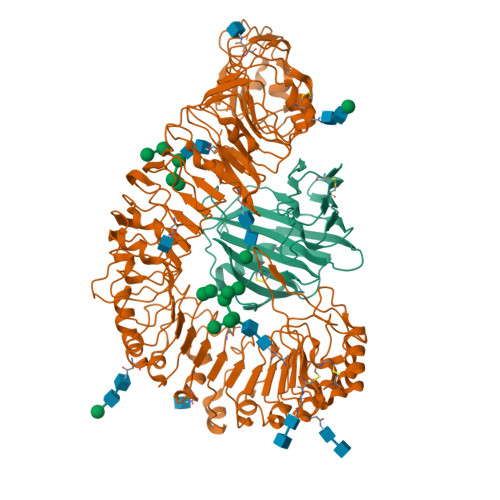
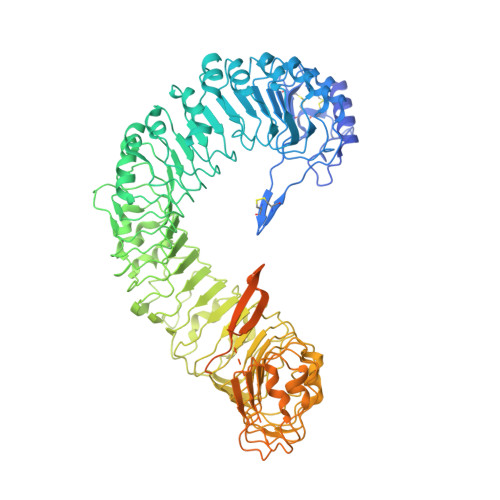
Plant receptor-like protein activation by a microbial glycoside hydrolase.
Sun, Y., Wang, Y., Zhang, X., Chen, Z., Xia, Y., Wang, L., Sun, Y., Zhang, M., Xiao, Y., Han, Z., Wang, Y., Chai, J.(2022) Nature 610: 335-342
- PubMed: 36131021
- DOI: https://doi.org/10.1038/s41586-022-05214-x
- Primary Citation of Related Structures:
7DRB, 7DRC, 7W3T, 7W3V, 7W3X - PubMed Abstract:
Plants rely on cell-surface-localized pattern recognition receptors to detect pathogen- or host-derived danger signals and trigger an immune response 1-6 . Receptor-like proteins (RLPs) with a leucine-rich repeat (LRR) ectodomain constitute a subgroup of pattern recognition receptors and play a critical role in plant immunity 1-3 . Mechanisms underlying ligand recognition and activation of LRR-RLPs remain elusive. Here we report a crystal structure of the LRR-RLP RXEG1 from Nicotiana benthamiana that recognizes XEG1 xyloglucanase from the pathogen Phytophthora sojae. The structure reveals that specific XEG1 recognition is predominantly mediated by an amino-terminal and a carboxy-terminal loop-out region (RXEG1(ID)) of RXEG1. The two loops bind to the active-site groove of XEG1, inhibiting its enzymatic activity and suppressing Phytophthora infection of N. benthamiana. Binding of XEG1 promotes association of RXEG1(LRR) with the LRR-type co-receptor BAK1 through RXEG1(ID) and the last four conserved LRRs to trigger RXEG1-mediated immune responses. Comparison of the structures of apo-RXEG1(LRR), XEG1-RXEG1(LRR) and XEG1-BAK1-RXEG1(LRR) shows that binding of XEG1 induces conformational changes in the N-terminal region of RXEG1(ID) and enhances structural flexibility of the BAK1-associating regions of RXEG1(LRR). These changes allow fold switching of RXEG1(ID) for recruitment of BAK1(LRR). Our data reveal a conserved mechanism of ligand-induced heterodimerization of an LRR-RLP with BAK1 and suggest a dual function for the LRR-RLP in plant immunity.
Organizational Affiliation:
Tsinghua-Peking Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, Center for Plant Biology, School of Life Sciences, Tsinghua University, Beijing, China.