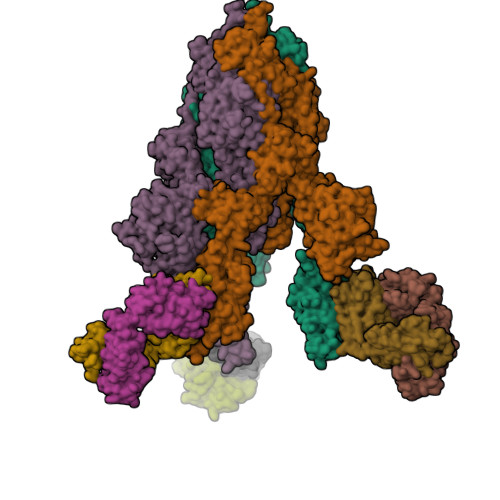
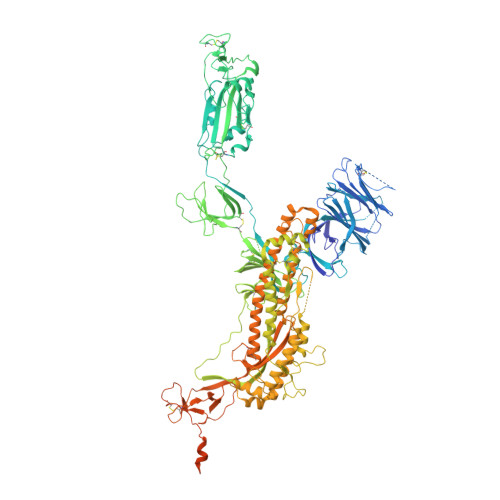
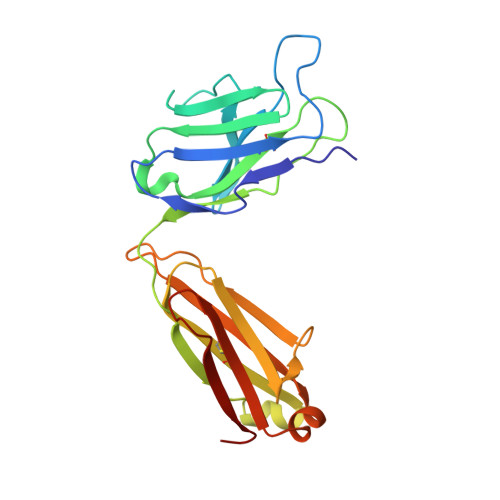
A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Wang, X., Hu, A., Chen, X., Zhang, Y., Yu, F., Yue, S., Li, A., Zhang, J., Pan, Z., Yang, Y., Lin, Y., Gao, L., Zhou, J., Zhao, J., Li, F., Shi, Y., Huang, F., Yang, X., Peng, Y., Tu, L., Zhang, H., Zheng, H., He, J., Zhang, H., Xu, L., Huang, Q., Zhu, Y., Deng, K., Ye, L.(2022) Signal Transduct Target Ther 7: 114