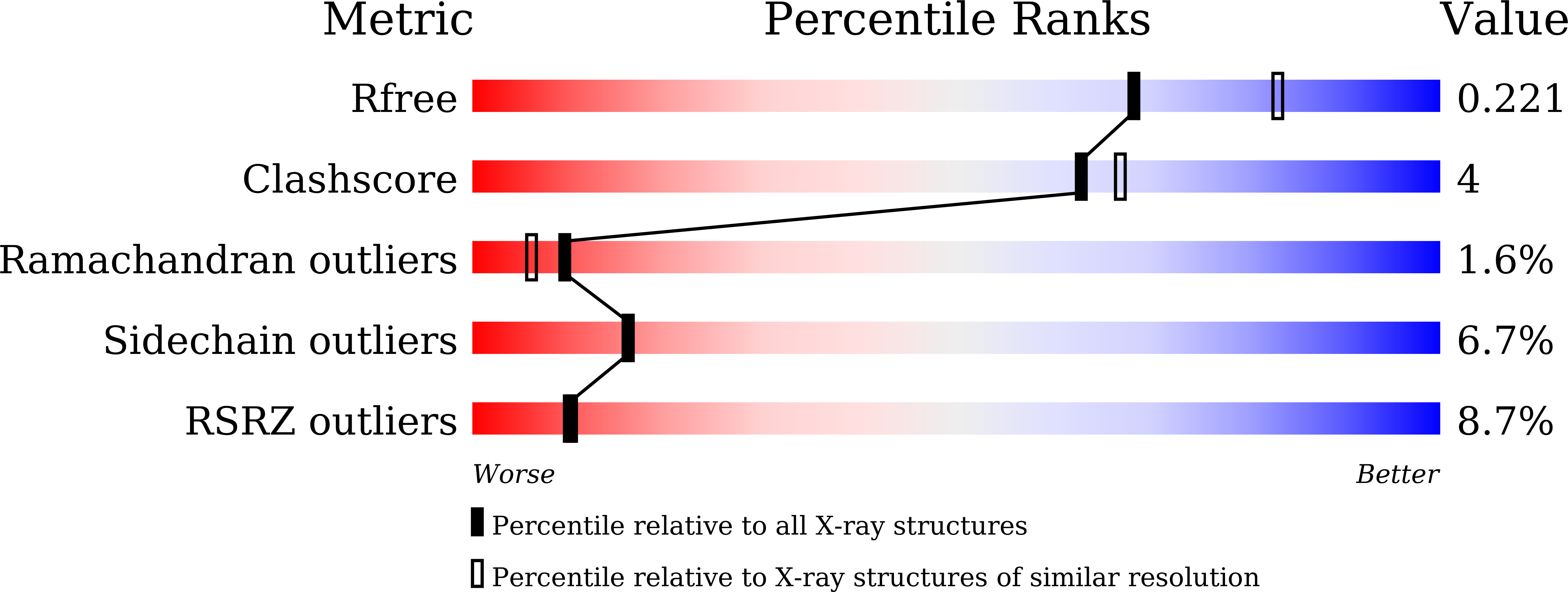
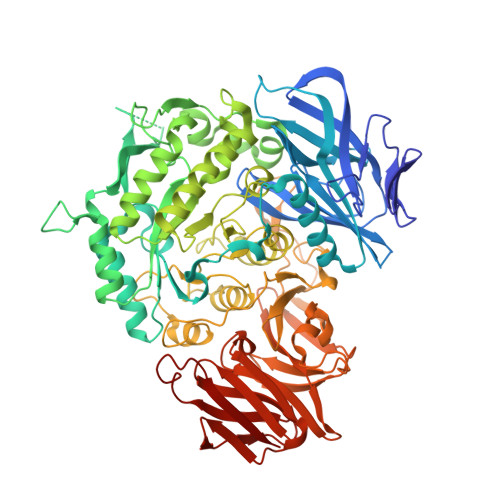
Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Chuankhayan, P., Lee, R.H., Guan, H.H., Lin, C.C., Chen, N.C., Huang, Y.C., Yoshimura, M., Nakagawa, A., Chen, C.J.(2023) Acta Crystallogr D Struct Biol 79: 154-167
- PubMed: 36762861
- DOI: https://doi.org/10.1107/S2059798323000037
- Primary Citation of Related Structures:
7EXF, 7EXG, 7EXH, 7EXJ, 7EXQ, 7EXR - PubMed Abstract:
The alkaline α-galactosidase AtAkαGal3 from Arabidopsis thaliana catalyzes the hydrolysis of α-D-galactose from galacto-oligosaccharides under alkaline conditions. A phylogenetic analysis based on sequence alignment classifies AtAkαGal3 as more closely related to the raffinose family of oligosaccharide (RFO) synthases than to the acidic α-galactosidases. Here, thin-layer chromatography is used to demonstrate that AtAkαGal3 exhibits a dual function and is capable of synthesizing stachyose using raffinose, instead of galactinol, as the galactose donor. Crystal structures of complexes of AtAkαGal3 and its D383A mutant with various substrates and products, including galactose, galactinol, raffinose, stachyose and sucrose, are reported as the first representative structures of an alkaline α-galactosidase. The structure of AtAkαGal3 comprises three domains: an N-terminal domain with 13 antiparallel β-strands, a catalytic domain with an (α/β) 8 -barrel fold and a C-terminal domain composed of β-sheets that form two Greek-key motifs. The WW box of the N-terminal domain, which comprises the conserved residues FRSK 75 XW 77 W 78 in the RFO synthases, contributes Trp77 and Trp78 to the +1 subsite to contribute to the substrate-binding ability together with the (α/β) 8 barrel of the catalytic domain. The C-terminal domain is presumably involved in structural stability. Structures of the D383A mutant in complex with various substrates and products, especially the natural substrate/product stachyose, reveal four complete subsites (-1 to +3) at the catalytic site. A functional loop (residues 329-352) that exists in the alkaline α-galactosidase AtAkαGal3 and possibly in RFO synthases, but not in acidic α-galactosidases, stabilizes the stachyose at the +2 and +3 subsites and extends the catalytic pocket for the transferase mechanism. Considering the similarities in amino-acid sequence, catalytic domain and activity between alkaline α-galactosidases and RFO synthases, the structure of AtAkαGal3 might also serve a model for the study of RFO synthases, structures of which are lacking.
Organizational Affiliation:
Life Science Group, Scientific Research Division, National Synchrotron Radiation Research Cente, Hsinchu 30076, Taiwan.