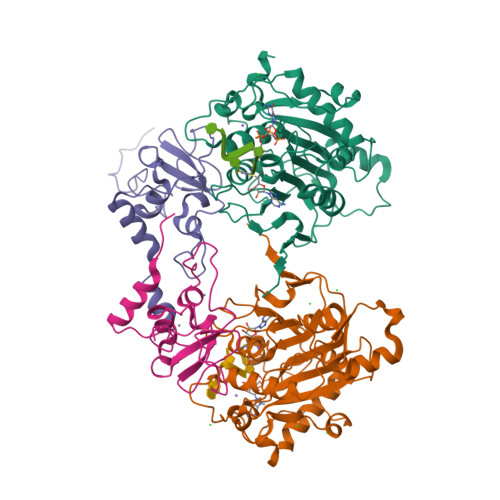
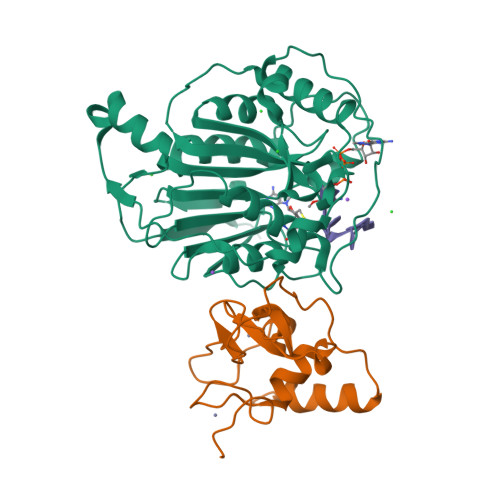
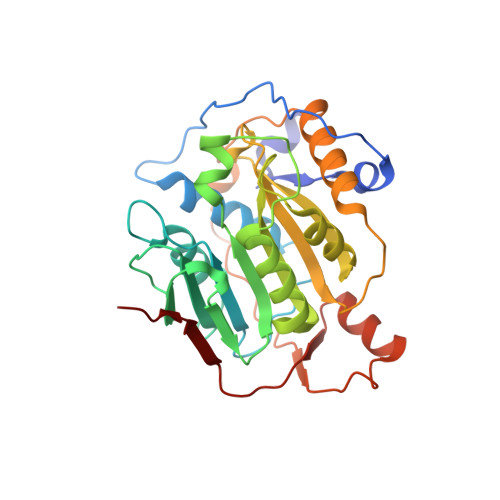
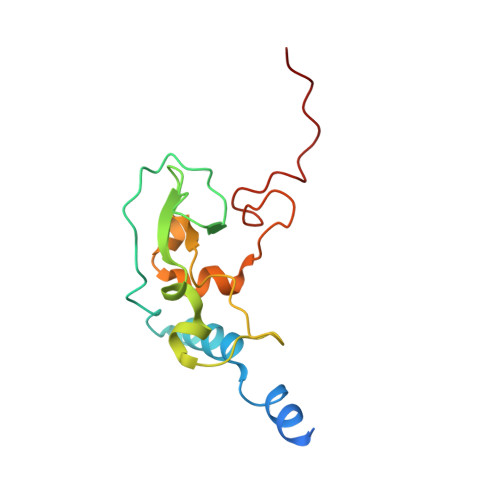
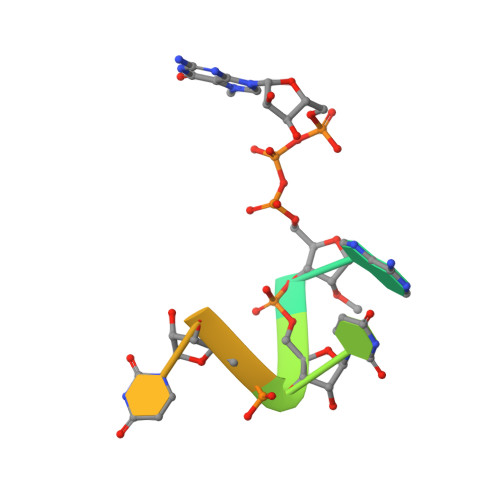
Structure of SARS-CoV-2 2'-O-methyltransferase heterodimer with RNA Cap analog and sulfates bound reveals new strategies for structure-based inhibitor design
Rosas-Lemus, M., Minasov, G., Shuvalova, L., Inniss, N., Kiryukhina, O., Brunzelle, J., Satchell, K.J.F.(2020) bioRxiv