Cryo-EM reveals the dynamic interplay between mitochondrial Hsp90 and SdhB folding intermediates
Liu, Y.X., Agard, D.A., Elnatan, D., Sun, M., Myasnikov, A.G.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Heat shock protein 75 kDa, mitochondrial | 651 | Homo sapiens | Mutation(s): 0 Gene Names: TRAP1, HSP75 | 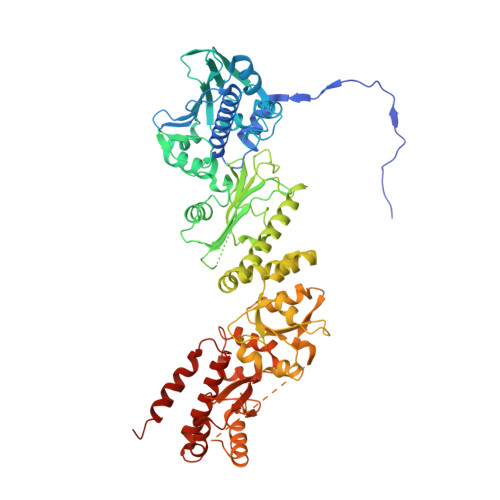 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q12931 (Homo sapiens) Explore Q12931 Go to UniProtKB: Q12931 | |||||
PHAROS: Q12931 GTEx: ENSG00000126602 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q12931 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ANP Query on ANP | E [auth A], H [auth B] | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER C10 H17 N6 O12 P3 PVKSNHVPLWYQGJ-KQYNXXCUSA-N |  | ||
| K Query on K | D [auth A], G [auth B] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| MG Query on MG | C [auth A], F [auth B] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Howard Hughes Medical Institute (HHMI) | United States | -- |
| American Heart Association | United States | 18POST33990362 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | R35GM118099 |
| National Institutes of Health/National Cancer Institute (NIH/NCI) | United States | U54CA209891 |
| National Institutes of Health/National Institute of Mental Health (NIH/NIMH) | United States | U01MH115747 |
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | U19AI135990 |