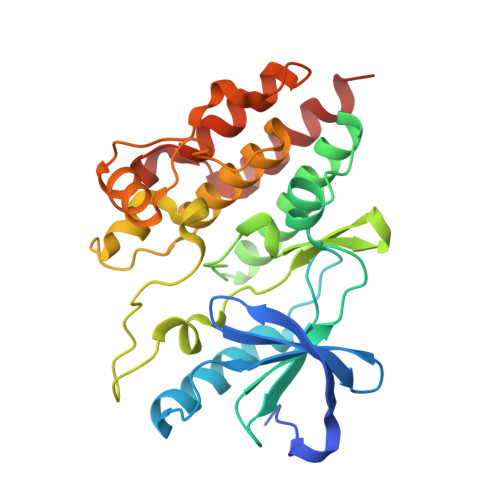
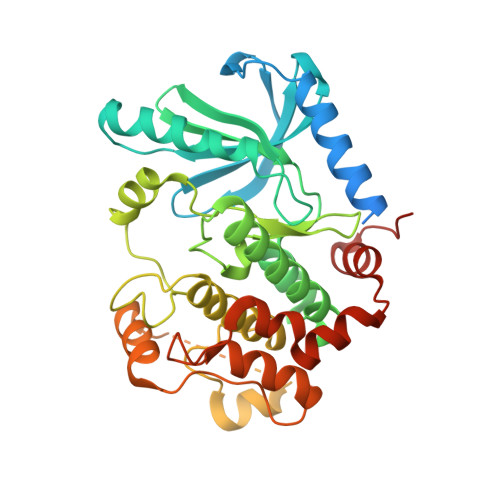
Allosteric MEK inhibitors act on BRAF/MEK complexes to block MEK activation.
Gonzalez-Del Pino, G.L., Li, K., Park, E., Schmoker, A.M., Ha, B.H., Eck, M.J.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34470822
- DOI: https://doi.org/10.1073/pnas.2107207118
- Primary Citation of Related Structures:
6V2W, 7M0T, 7M0U, 7M0V, 7M0W, 7M0X, 7M0Y, 7M0Z - PubMed Abstract:
The RAF/MEK/ERK pathway is central to the control of cell physiology, and its dysregulation is associated with many cancers. Accordingly, the proteins constituting this pathway, including MEK1/2 (MEK), have been subject to intense drug discovery and development efforts. Allosteric MEK inhibitors (MEKi) exert complex effects on RAF/MEK/ERK pathway signaling and are employed clinically in combination with BRAF inhibitors in malignant melanoma. Although mechanisms and structures of MEKi bound to MEK have been described for many of these compounds, recent studies suggest that RAF/MEK complexes, rather than free MEK, should be evaluated as the target of MEKi. Here, we describe structural and biochemical studies of eight structurally diverse, clinical-stage MEKi to better understand their mechanism of action on BRAF/MEK complexes. We find that all of these agents bind in the MEK allosteric site in BRAF/MEK complexes, in which they stabilize the MEK activation loop in a conformation that is resistant to BRAF-mediated dual phosphorylation required for full activation of MEK. We also show that allosteric MEK inhibitors act most potently on BRAF/MEK complexes rather than on free active MEK, further supporting the notion that a BRAF/MEK complex is the physiologically relevant pharmacologic target for this class of compounds. Our findings provide a conceptual and structural framework for rational development of RAF-selective MEK inhibitors as an avenue to more effective and better-tolerated agents targeting this pathway.
Organizational Affiliation:
Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA 02215.