Structural and functional insight into mismatch extension by human DNA polymerase alpha.
Baranovskiy, A.G., Babayeva, N.D., Lisova, A.E., Morstadt, L.M., Tahirov, T.H.(2022) Proc Natl Acad Sci U S A 119: e2111744119-e2111744119
- PubMed: 35467978
- DOI: https://doi.org/10.1073/pnas.2111744119
- Primary Citation of Related Structures:
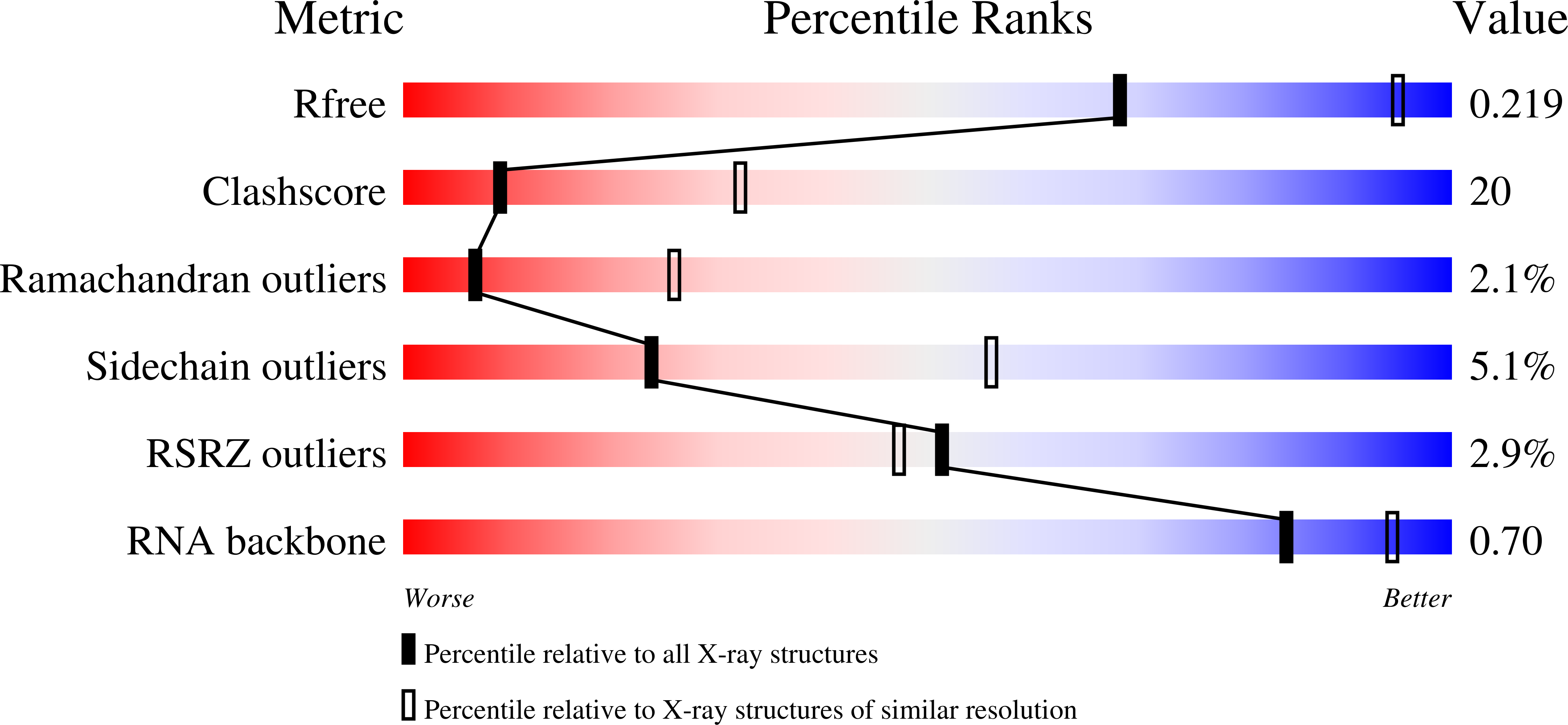
7N2M - PubMed Abstract:
Human DNA polymerase α (Polα) does not possess proofreading ability and plays an important role in genome replication and mutagenesis. Polα extends the RNA primers generated by primase and provides a springboard for loading other replication factors. Here we provide the structural and functional analysis of the human Polα interaction with a mismatched template:primer. The structure of the human Polα catalytic domain in the complex with an incoming deoxycytidine triphosphate (dCTP) and the template:primer containing a T-C mismatch at the growing primer terminus was solved at a 2.9 Å resolution. It revealed the absence of significant distortions in the active site and in the conformation of the substrates, except the primer 3′-end. The T-C mismatch acquired a planar geometry where both nucleotides moved toward each other by 0.4 Å and 0.7 Å, respectively, and made one hydrogen bond. The binding studies conducted at a physiological salt concentration revealed that Polα has a low affinity to DNA and is not able to discriminate against a mispaired template:primer in the absence of deoxynucleotide triphosphate (dNTP). Strikingly, in the presence of cognate dNTP, Polα showed a more than 10-fold higher selectivity for a correct duplex versus a mismatched one. According to pre-steady-state kinetic studies, human Polα extends the T-C mismatch with a 249-fold lower efficiency due to reduction of the polymerization rate constant by 38-fold and reduced affinity to the incoming nucleotide by 6.6-fold. Thus, a mismatch at the postinsertion site affects all factors important for primer extension: affinity to both substrates and the rate of DNA polymerization.
Organizational Affiliation:
Eppley Institute for Research in Cancer and Allied Diseases, Fred & Pamela Buffett Cancer Center, University of Nebraska Medical Center, Omaha, NE 68198.