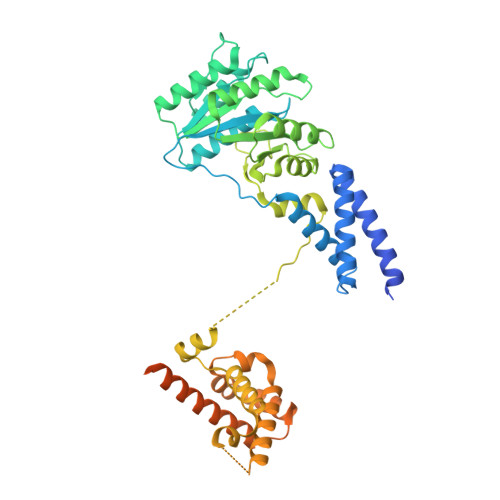
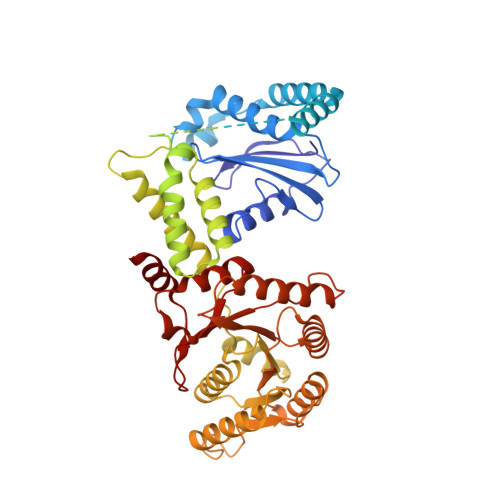
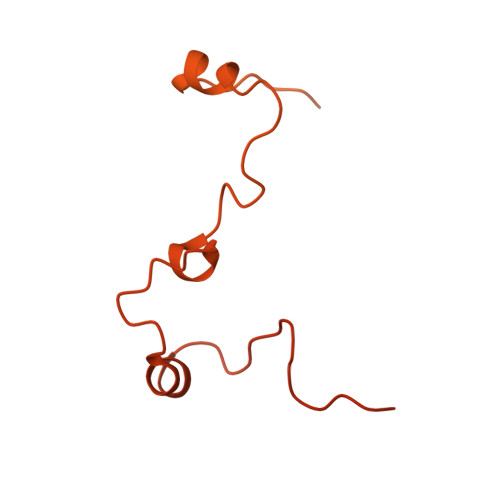
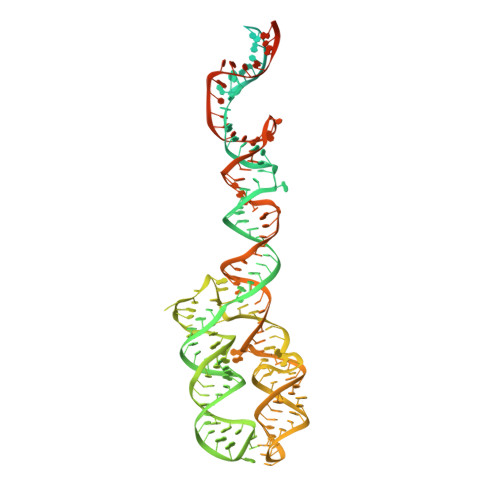
Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle.
Jomaa, A., Eitzinger, S., Zhu, Z., Chandrasekar, S., Kobayashi, K., Shan, S.O., Ban, N.(2021) Cell Rep 36: 109350-109350
- PubMed: 34260909
- DOI: https://doi.org/10.1016/j.celrep.2021.109350
- Primary Citation of Related Structures:
7OBQ, 7OBR - PubMed Abstract:
Co-translational protein targeting to membranes by the signal recognition particle (SRP) is a universally conserved pathway from bacteria to humans. In mammals, SRP and its receptor (SR) have many additional RNA features and protein components compared to the bacterial system, which were recently shown to play regulatory roles. Due to its complexity, the mammalian SRP targeting process is mechanistically not well understood. In particular, it is not clear how SRP recognizes translating ribosomes with exposed signal sequences and how the GTPase activity of SRP and SR is regulated. Here, we present electron cryo-microscopy structures of SRP and SRP·SR in complex with the translating ribosome. The structures reveal the specific molecular interactions between SRP and the emerging signal sequence and the elements that regulate GTPase activity of SRP·SR. Our results suggest the molecular mechanism of how eukaryote-specific elements regulate the early and late stages of SRP-dependent protein targeting.
Organizational Affiliation:
Department of Biology, Institute of Molecular Biology and Biophysics, ETH Zurich, 8093 Zurich, Switzerland. Electronic address: ahmad.jomaa@mol.biol.ethz.ch.