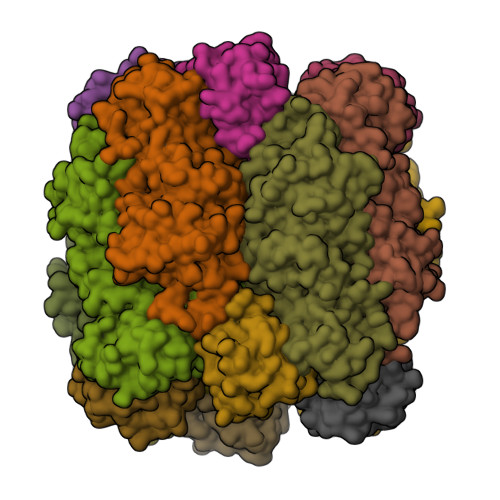
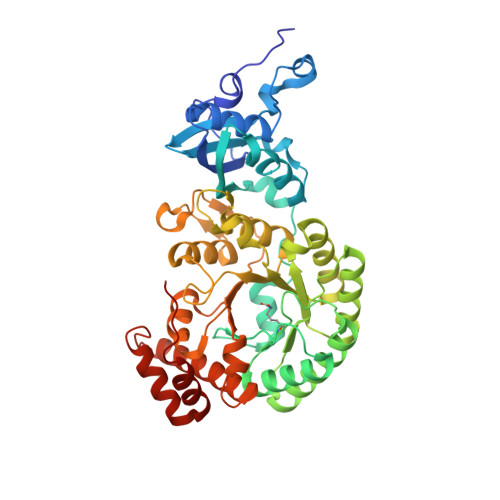
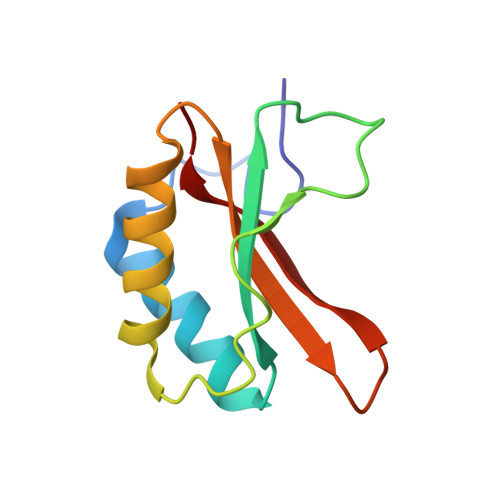
Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Schulz, L., Guo, Z., Zarzycki, J., Steinchen, W., Schuller, J.M., Heimerl, T., Prinz, S., Mueller-Cajar, O., Erb, T.J., Hochberg, G.K.A.(2022) Science 378: 155-160
- PubMed: 36227987
- DOI: https://doi.org/10.1126/science.abq1416
- Primary Citation of Related Structures:
7QSV, 7QSW, 7QSX, 7QSY, 7QSZ, 7QT1, 7QVI - PubMed Abstract:
The evolution of ribulose-1,5-bisphosphate carboxylase/oxygenases (Rubiscos) that discriminate strongly between their substrate carbon dioxide and the undesired side substrate dioxygen was an important event for photosynthetic organisms adapting to an oxygenated environment. We use ancestral sequence reconstruction to recapitulate this event. We show that Rubisco increased its specificity and carboxylation efficiency through the gain of an accessory subunit before atmospheric oxygen was present. Using structural and biochemical approaches, we retrace how this subunit was gained and became essential. Our work illuminates the emergence of an adaptation to rising ambient oxygen levels, provides a template for investigating the function of interactions that have remained elusive because of their essentiality, and sheds light on the determinants of specificity in Rubisco.
Organizational Affiliation:
Department of Biochemistry and Synthetic Metabolism, Max Planck Institute for Terrestrial Microbiology, 35043 Marburg, Germany.