Patient-derived monoclonal antibody neutralizes SARS-CoV-2 Omicron variants and confers full protection in monkeys.
Fenwick, C., Turelli, P., Ni, D., Perez, L., Lau, K., Herate, C., Marlin, R., Lana, E., Pellaton, C., Raclot, C., Esteves-Leuenberger, L., Campos, J., Farina, A., Fiscalini, F., Dereuddre-Bosquet, N., Relouzat, F., Abdelnabi, R., Foo, C.S., Neyts, J., Leyssen, P., Levy, Y., Pojer, F., Stahlberg, H., LeGrand, R., Trono, D., Pantaleo, G.(2022) Nat Microbiol 7: 1376-1389
- PubMed: 35879526
- DOI: https://doi.org/10.1038/s41564-022-01198-6
- Primary Citation of Related Structures:
7QTI, 7QTJ, 7QTK - PubMed Abstract:
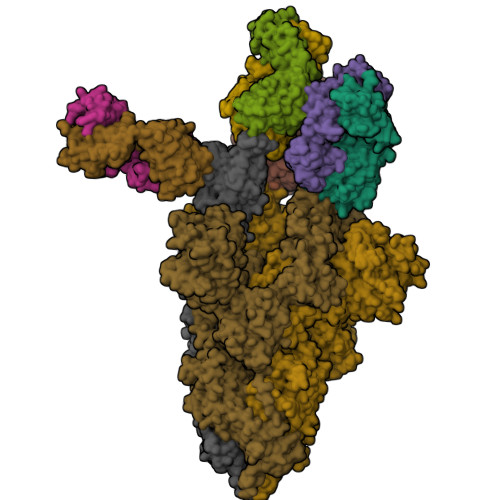
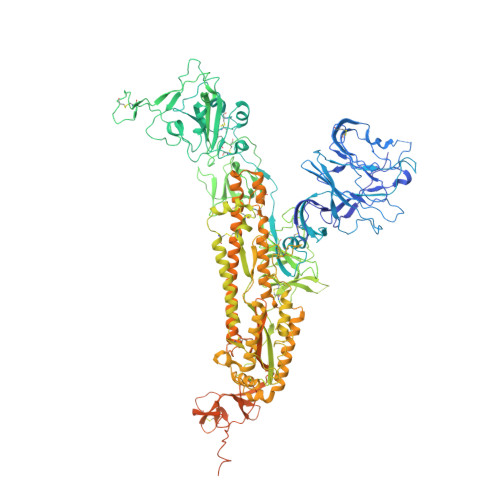
The SARS-CoV-2 Omicron variant has very high levels of transmission, is resistant to neutralization by authorized therapeutic human monoclonal antibodies (mAb) and is less sensitive to vaccine-mediated immunity. To provide additional therapies against Omicron, we isolated a mAb named P2G3 from a previously infected vaccinated donor and showed that it has picomolar-range neutralizing activity against Omicron BA.1, BA.1.1, BA.2 and all other variants tested. We solved the structure of P2G3 Fab in complex with the Omicron spike using cryo-electron microscopy at 3.04 Å resolution to identify the P2G3 epitope as a Class 3 mAb that is different from mAb-binding spike epitopes reported previously. Using a SARS-CoV-2 Omicron monkey challenge model, we show that P2G3 alone, or in combination with P5C3 (a broadly active Class 1 mAb previously identified), confers complete prophylactic or therapeutic protection. Although we could select for SARS-CoV-2 mutants escaping neutralization by P2G3 or by P5C3 in vitro, they had low infectivity and 'escape' mutations are extremely rare in public sequence databases. We conclude that this combination of mAbs has potential as an anti-Omicron drug.
Organizational Affiliation:
Service of Immunology and Allergy, Department of Medicine, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland.