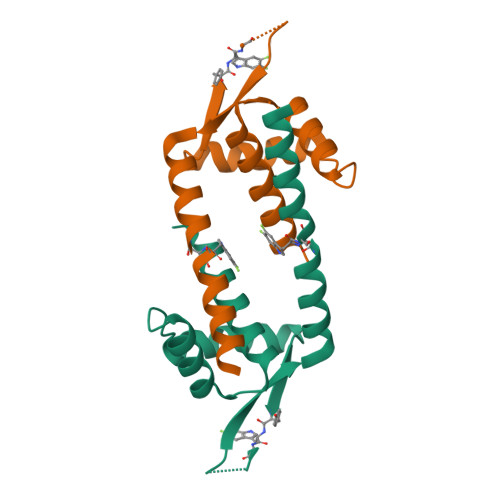
The Role of Tryptophan in pi Interactions in Proteins: An Experimental Approach.
Shao, J., Kuiper, B.P., Thunnissen, A.W.H., Cool, R.H., Zhou, L., Huang, C., Dijkstra, B.W., Broos, J.(2022) J Am Chem Soc 144: 13815-13822
- PubMed: 35868012
- DOI: https://doi.org/10.1021/jacs.2c04986
- Primary Citation of Related Structures:
7QZ5, 7QZ6, 7QZ7, 7QZ8, 7QZ9 - PubMed Abstract:
In proteins, the amino acids Phe, Tyr, and especially Trp are frequently involved in π interactions such as π-π, cation-π, and CH-π bonds. These interactions are often crucial for protein structure and protein-ligand binding. A powerful means to study these interactions is progressive fluorination of these aromatic residues to modulate the electrostatic component of the interaction. However, to date no protein expression platform is available to produce milligram amounts of proteins labeled with such fluorinated amino acids. Here, we present a Lactococcus lactis Trp auxotroph-based expression system for efficient incorporation (≥95%) of mono-, di-, tri-, and tetrafluorinated, as well as a methylated Trp analog. As a model protein we have chosen LmrR, a dimeric multidrug transcriptional repressor protein from L. lactis. LmrR binds aromatic drugs, like daunomycin and riboflavin, between Trp96 and Trp96' in the dimer interface. Progressive fluorination of Trp96 decreased the affinity for the drugs 6- to 70-fold, clearly establishing the importance of electrostatic π-π interactions for drug binding. Presteady state kinetic data of the LmrR-drug interaction support the enthalpic nature of the interaction, while high resolution crystal structures of the labeled protein-drug complexes provide for the first time a structural view of the progressive fluorination approach. The L. lactis expression system was also used to study the role of Trp68 in the binding of riboflavin by the membrane-bound riboflavin transport protein RibU from L. lactis . Progressive fluorination of Trp68 revealed a strong electrostatic component that contributed 15-20% to the total riboflavin-RibU binding energy.
Organizational Affiliation:
Groningen Biomolecular Science and Biotechnology Institute (GBB), University of Groningen, Nijenborgh 7, 9747 AG Groningen, The Netherlands.