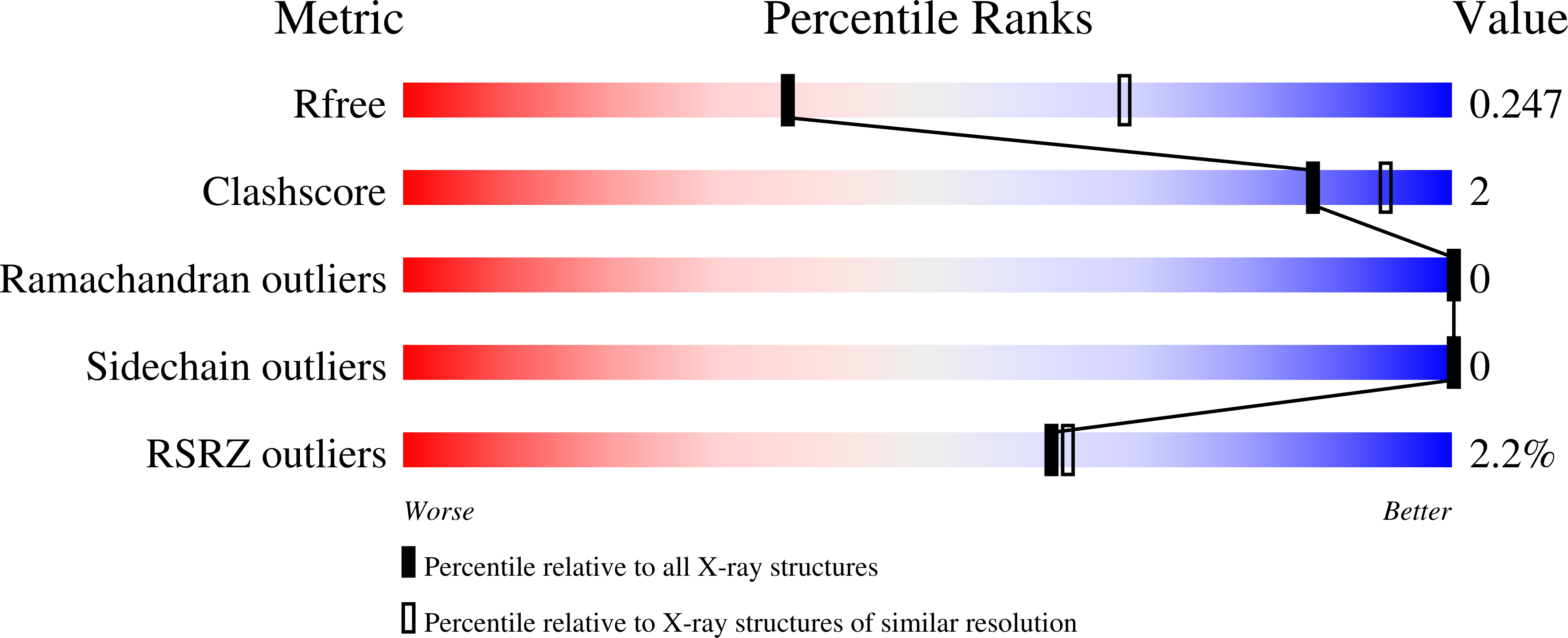
Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Klima, M., Khalili Yazdi, A., Li, F., Chau, I., Hajian, T., Bolotokova, A., Kaniskan, H.U., Han, Y., Wang, K., Li, D., Luo, M., Jin, J., Boura, E., Vedadi, M.(2022) Protein Sci 31: e4395-e4395
- PubMed: 36040262
- DOI: https://doi.org/10.1002/pro.4395
- Primary Citation of Related Structures:
7R1T, 7R1U - PubMed Abstract:
SARS-CoV-2 nsp10-nsp16 complex is a 2'-O-methyltransferase (MTase) involved in viral RNA capping, enabling the virus to evade the immune system in humans. It has been considered a valuable target in the discovery of antiviral therapeutics, as the RNA cap formation is crucial for viral propagation. Through cross-screening of the inhibitors that we previously reported for SARS-CoV-2 nsp14 MTase activity against nsp10-nsp16 complex, we identified two compounds (SS148 and WZ16) that also inhibited nsp16 MTase activity. To further enable the chemical optimization of these two compounds towards more potent and selective dual nsp14/nsp16 MTase inhibitors, we determined the crystal structure of nsp10-nsp16 in complex with each of SS148 and WZ16. As expected, the structures revealed the binding of both compounds to S-adenosyl-L-methionine (SAM) binding pocket of nsp16. However, our structural data along with the biochemical mechanism of action determination revealed an RNA-dependent SAM-competitive pattern of inhibition for WZ16, clearly suggesting that binding of the RNA first may help the binding of some SAM competitive inhibitors. Both compounds also showed some degree of selectivity against human protein MTases, an indication of great potential for chemical optimization towards more potent and selective inhibitors of coronavirus MTases.
Organizational Affiliation:
Institute of Organic Chemistry and Biochemistry, Czech Academy of Sciences, Prague 6, Czech Republic.