Peptidomimetic nitrile warheads as SARS-CoV-2 3CL protease inhibitors.
Bai, B., Arutyunova, E., Khan, M.B., Lu, J., Joyce, M.A., Saffran, H.A., Shields, J.A., Kandadai, A.S., Belovodskiy, A., Hena, M., Vuong, W., Lamer, T., Young, H.S., Vederas, J.C., Tyrrell, D.L., Lemieux, M.J., Nieman, J.A.(2021) RSC Med Chem 12: 1722-1730
- PubMed: 34778773
- DOI: https://doi.org/10.1039/d1md00247c
- Primary Citation of Related Structures:
7R7H - PubMed Abstract:
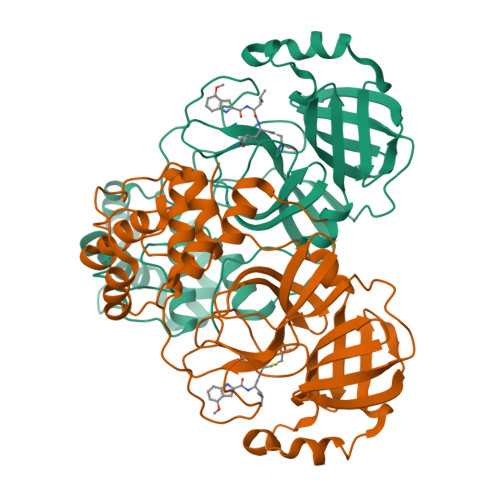
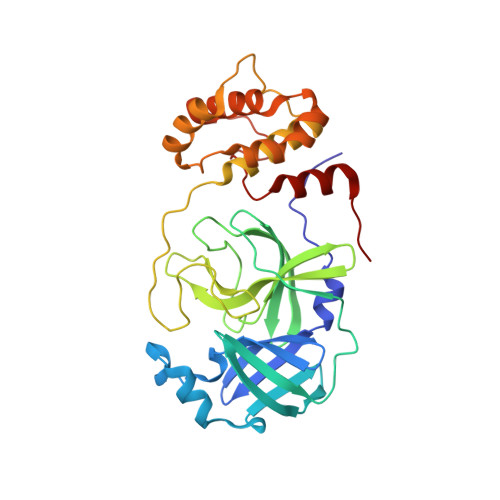
Tragically, the death toll from the COVID-19 pandemic continues to rise, and with variants being observed around the globe new therapeutics, particularly direct-acting antivirals that are easily administered, are desperately needed. Studies targeting the SARS-CoV-2 3CL protease, which is critical for viral replication, with different peptidomimetics and warheads is an active area of research for development of potential drugs. To date, however, only a few publications have evaluated the nitrile warhead as a viral 3CL protease inhibitor, with only modest activity reported. This article describes our investigation of P3 4-methoxyindole peptidomimetic analogs with select P1 and P2 groups with a nitrile warhead that are potent inhibitors of SARS-CoV-2 3CL protease and demonstrate in vitro SARS-CoV-2 antiviral activity. A selectivity for SARS-CoV-2 3CL protease over human cathepsins B, S and L was also observed with the nitrile warhead, which was superior to that with the aldehyde warhead. A co-crystal structure with SARS-CoV-2 3CL protease and a reversibility study indicate that a reversible, thioimidate adduct is formed when the catalytic sulfur forms a covalent bond with the carbon of the nitrile. This effort also identified efflux as a property limiting antiviral activity of these compounds, and together with the positive attributes described these results provide insight for further drug development of novel nitrile peptidomimetics targeting SARS-CoV-2 3CL protease.
Organizational Affiliation:
Li Ka Shing Applied Virology Institute, University of Alberta Edmonton Alberta T6G 2E1 Canada jnieman@ualberta.ca.