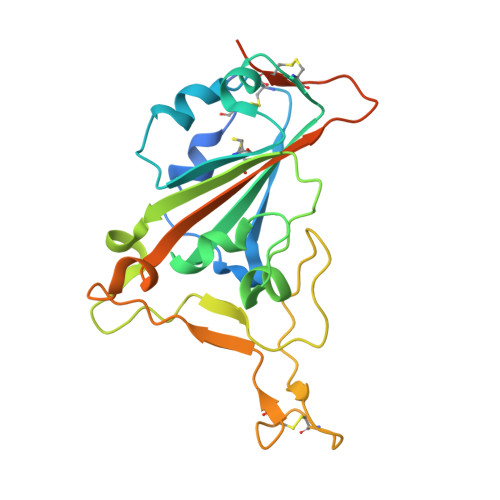
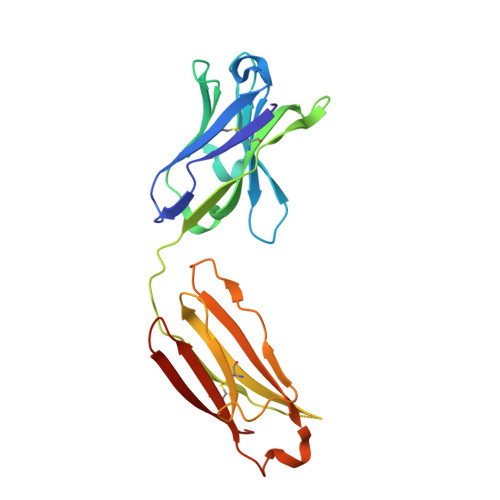
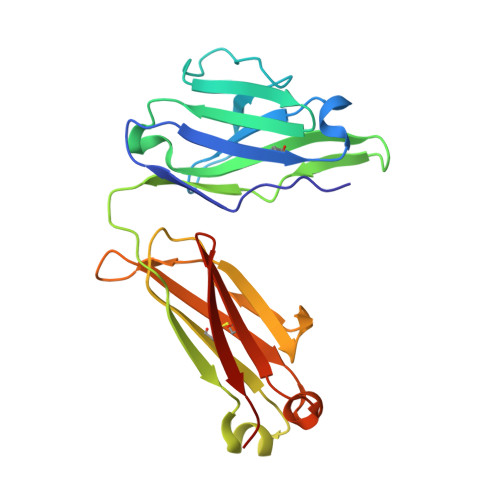
SARS-CoV-2 Beta variant infection elicits potent lineage-specific and cross-reactive antibodies.
Reincke, S.M., Yuan, M., Kornau, H.C., Corman, V.M., van Hoof, S., Sanchez-Sendin, E., Ramberger, M., Yu, W., Hua, Y., Tien, H., Schmidt, M.L., Schwarz, T., Jeworowski, L.M., Brandl, S.E., Rasmussen, H.F., Homeyer, M.A., Stoffler, L., Barner, M., Kunkel, D., Huo, S., Horler, J., von Wardenburg, N., Kroidl, I., Eser, T.M., Wieser, A., Geldmacher, C., Hoelscher, M., Ganzer, H., Weiss, G., Schmitz, D., Drosten, C., Pruss, H., Wilson, I.A., Kreye, J.(2022) Science 375: 782-787
- PubMed: 35076281
- DOI: https://doi.org/10.1126/science.abm5835
- Primary Citation of Related Structures:
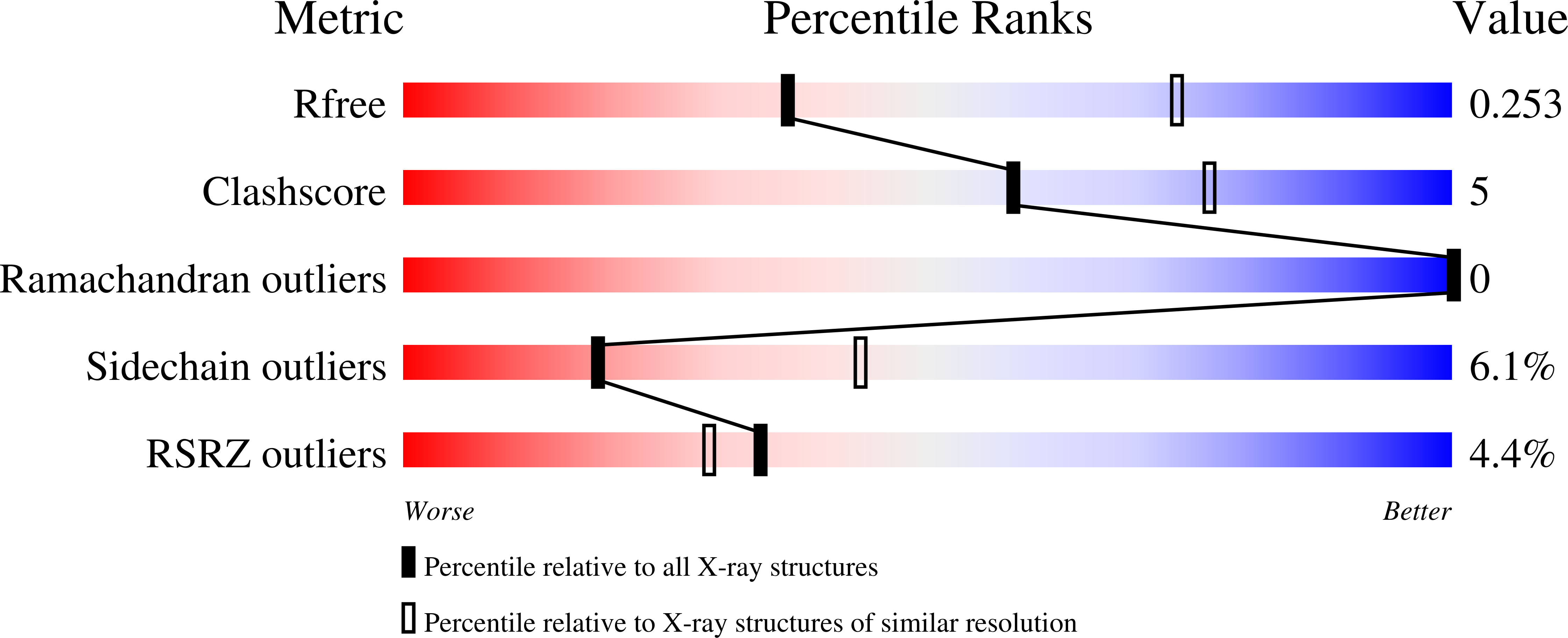
7S5P, 7S5Q, 7S5R - PubMed Abstract:
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Beta variant of concern (VOC) resists neutralization by major classes of antibodies from COVID-19 patients and vaccinated individuals. In this study, serum of Beta-infected patients revealed reduced cross-neutralization of wild-type virus. From these patients, we isolated Beta-specific and cross-reactive receptor-binding domain (RBD) antibodies. The Beta-specificity results from recruitment of VOC-specific clonotypes and accommodation of mutations present in Beta and Omicron into a major antibody class that is normally sensitive to these mutations. The Beta-elicited cross-reactive antibodies share genetic and structural features with wild type-elicited antibodies, including a public VH1-58 clonotype that targets the RBD ridge. These findings advance our understanding of the antibody response to SARS-CoV-2 shaped by antigenic drift, with implications for design of next-generation vaccines and therapeutics.
Organizational Affiliation:
Department of Neurology and Experimental Neurology, Charité-Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Berlin, Germany.