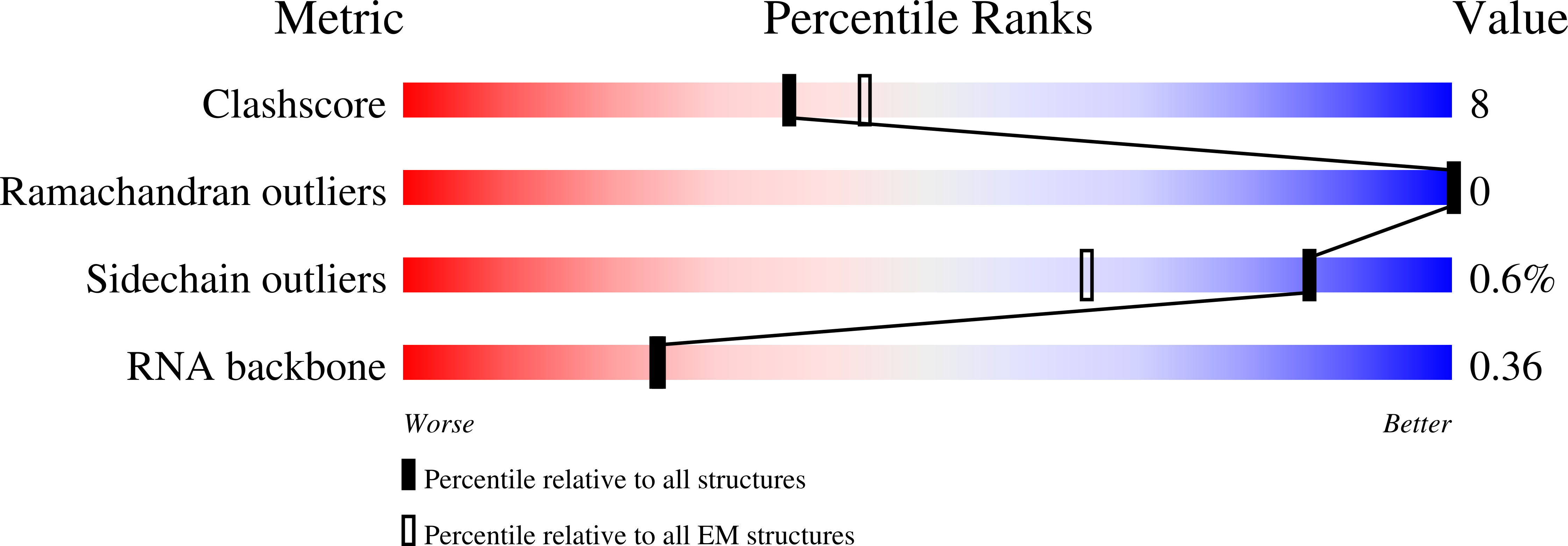
Structural basis for RNA slicing by a plant Argonaute.
Xiao, Y., Maeda, S., Otomo, T., MacRae, I.J.(2023) Nat Struct Mol Biol 30: 778-784
- PubMed: 37127820
- DOI: https://doi.org/10.1038/s41594-023-00989-7
- Primary Citation of Related Structures:
7SVA, 7SWQ - PubMed Abstract:
Argonaute (AGO) proteins use small RNAs to recognize transcripts targeted for silencing in plants and animals. Many AGOs cleave target RNAs using an endoribonuclease activity termed 'slicing'. Slicing by DNA-guided prokaryotic AGOs has been studied in detail, but structural insights into RNA-guided slicing by eukaryotic AGOs are lacking. Here we present cryogenic electron microscopy structures of the Arabidopsis thaliana Argonaute10 (AtAgo10)-guide RNA complex with and without a target RNA representing a slicing substrate. The AtAgo10-guide-target complex adopts slicing-competent and slicing-incompetent conformations that are unlike known prokaryotic AGO structures. AtAgo10 slicing activity is licensed by docking target (t) nucleotides t9-t13 into a surface channel containing the AGO endoribonuclease active site. A β-hairpin in the L1 domain secures the t9-t13 segment and coordinates t9-t13 docking with extended guide-target pairing. Results show that prokaryotic and eukaryotic AGOs use distinct mechanisms for achieving target slicing and provide insights into small interfering RNA potency.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA, USA.