Multivalent interactions essential for lentiviral integrase function.
Ballandras-Colas, A., Chivukula, V., Gruszka, D.T., Shan, Z., Singh, P.K., Pye, V.E., McLean, R.K., Bedwell, G.J., Li, W., Nans, A., Cook, N.J., Fadel, H.J., Poeschla, E.M., Griffiths, D.J., Vargas, J., Taylor, I.A., Lyumkis, D., Yardimci, H., Engelman, A.N., Cherepanov, P.(2022) Nat Commun 13: 2416-2416
- PubMed: 35504909
- DOI: https://doi.org/10.1038/s41467-022-29928-8
- Primary Citation of Related Structures:
7U32 - PubMed Abstract:
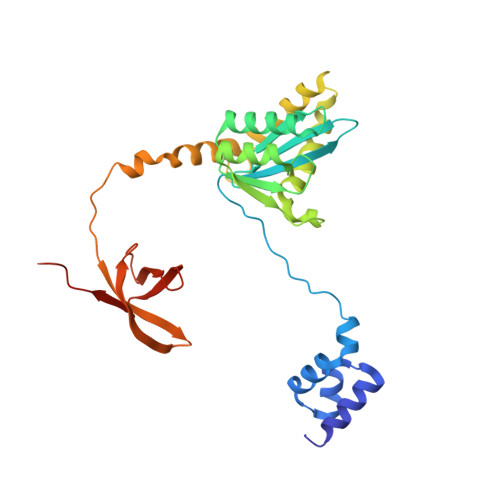
A multimer of retroviral integrase (IN) synapses viral DNA ends within a stable intasome nucleoprotein complex for integration into a host cell genome. Reconstitution of the intasome from the maedi-visna virus (MVV), an ovine lentivirus, revealed a large assembly containing sixteen IN subunits 1 . Herein, we report cryo-EM structures of the lentiviral intasome prior to engagement of target DNA and following strand transfer, refined at 3.4 and 3.5 Å resolution, respectively. The structures elucidate details of the protein-protein and protein-DNA interfaces involved in lentiviral intasome formation. We show that the homomeric interfaces involved in IN hexadecamer formation and the α-helical configuration of the linker connecting the C-terminal and catalytic core domains are critical for MVV IN strand transfer activity in vitro and for virus infectivity. Single-molecule microscopy in conjunction with photobleaching reveals that the MVV intasome can bind a variable number, up to sixteen molecules, of the lentivirus-specific host factor LEDGF/p75. Concordantly, ablation of endogenous LEDGF/p75 results in gross redistribution of MVV integration sites in human and ovine cells. Our data confirm the importance of the expanded architecture observed in cryo-EM studies of lentiviral intasomes and suggest that this organization underlies multivalent interactions with chromatin for integration targeting to active genes.
Organizational Affiliation:
Chromatin Structure and Mobile DNA Laboratory, The Francis Crick Institute, London, UK.