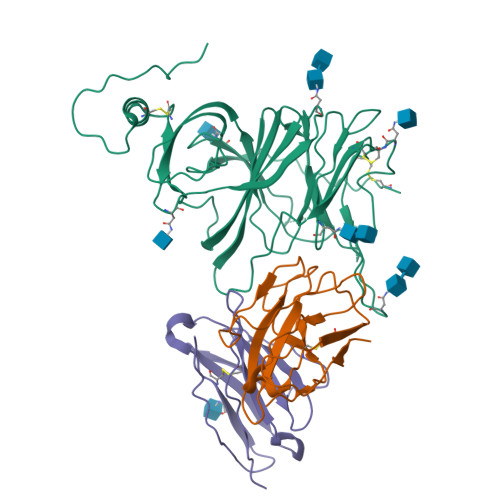
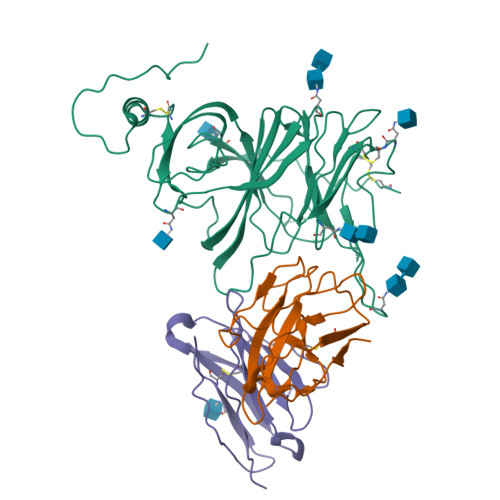
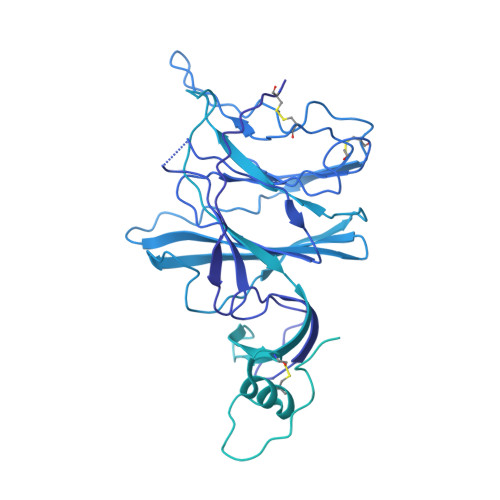
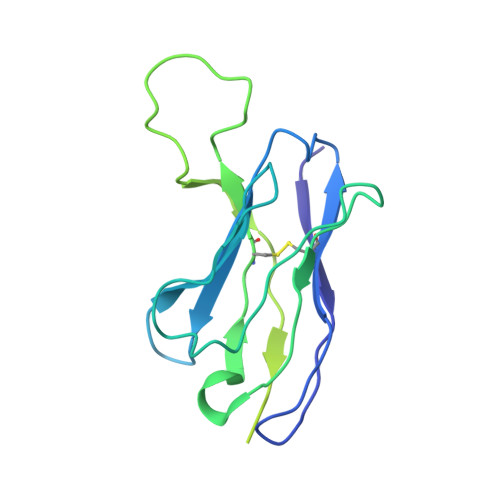
Analysis of memory B cells identifies conserved neutralizing epitopes on the N-terminal domain of variant SARS-Cov-2 spike proteins.
Wang, Z., Muecksch, F., Cho, A., Gaebler, C., Hoffmann, H.H., Ramos, V., Zong, S., Cipolla, M., Johnson, B., Schmidt, F., DaSilva, J., Bednarski, E., Ben Tanfous, T., Raspe, R., Yao, K., Lee, Y.E., Chen, T., Turroja, M., Milard, K.G., Dizon, J., Kaczynska, A., Gazumyan, A., Oliveira, T.Y., Rice, C.M., Caskey, M., Bieniasz, P.D., Hatziioannou, T., Barnes, C.O., Nussenzweig, M.C.(2022) Immunity 55: 998-1012.e8
- PubMed: 35447092
- DOI: https://doi.org/10.1016/j.immuni.2022.04.003
- Primary Citation of Related Structures:
7UAP, 7UAQ, 7UAR - PubMed Abstract:
SARS-CoV-2 infection or vaccination produces neutralizing antibody responses that contribute to better clinical outcomes. The receptor-binding domain (RBD) and the N-terminal domain (NTD) of the spike trimer (S) constitute the two major neutralizing targets for antibodies. Here, we use NTD-specific probes to capture anti-NTD memory B cells in a longitudinal cohort of infected individuals, some of whom were vaccinated. We found 6 complementation groups of neutralizing antibodies. 58% targeted epitopes outside the NTD supersite, 58% neutralized either Gamma or Omicron, and 14% were broad neutralizers that also neutralized Omicron. Structural characterization revealed that broadly active antibodies targeted three epitopes outside the NTD supersite including a class that recognized both the NTD and SD2 domain. Rapid recruitment of memory B cells producing these antibodies into the plasma cell compartment upon re-infection likely contributes to the relatively benign course of subsequent infections with SARS-CoV-2 variants, including Omicron.
Organizational Affiliation:
Laboratory of Molecular Immunology, The Rockefeller University, New York, NY 10065, USA.