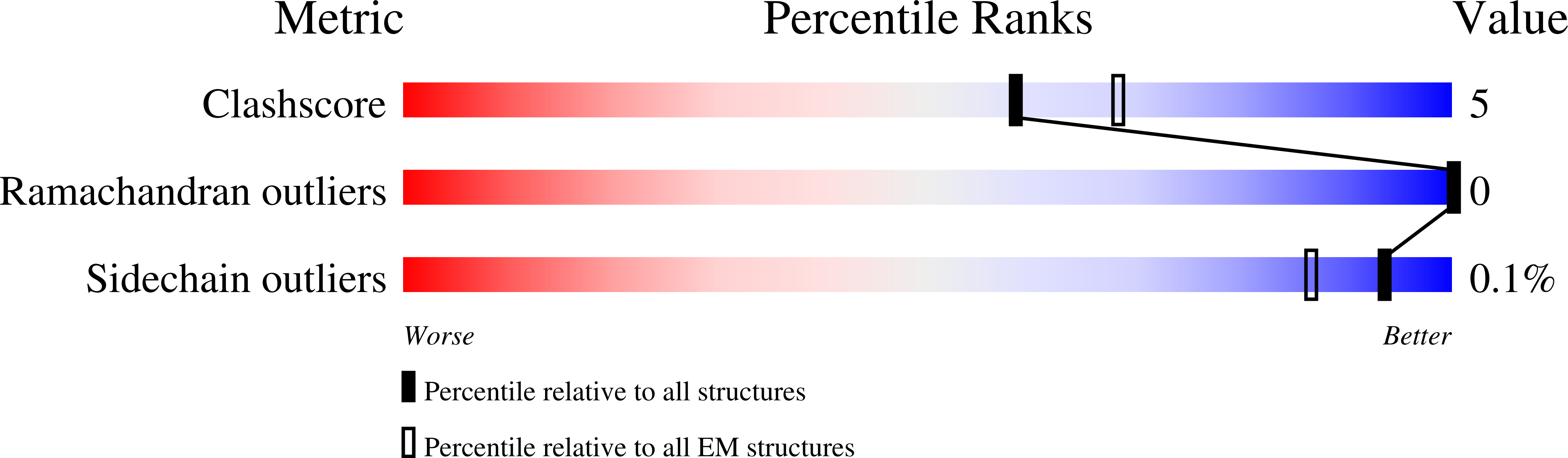Cryo-EM structures of SARS-CoV-2 Omicron BA.2 spike.
Stalls, V., Lindenberger, J., Gobeil, S.M., Henderson, R., Parks, R., Barr, M., Deyton, M., Martin, M., Janowska, K., Huang, X., May, A., Speakman, M., Beaudoin, E., Kraft, B., Lu, X., Edwards, R.J., Eaton, A., Montefiori, D.C., Williams, W.B., Saunders, K.O., Wiehe, K., Haynes, B.F., Acharya, P.(2022) Cell Rep 39: 111009-111009
- PubMed: 35732171
- DOI: https://doi.org/10.1016/j.celrep.2022.111009
- Primary Citation of Related Structures:
7UB0, 7UB5, 7UB6 - PubMed Abstract:
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron BA.2 sub-lineage has gained in proportion relative to BA.1. Because spike (S) protein variations may underlie differences in their pathobiology, here we determine cryoelectron microscopy (cryo-EM) structures of the BA.2 S ectodomain and compare these with previously determined BA.1 S structures. BA.2 receptor-binding domain (RBD) mutations induce remodeling of the RBD structure, resulting in tighter packing and improved thermostability. Interprotomer RBD interactions are enhanced in the closed (or 3-RBD-down) BA.2 S, while the fusion peptide is less accessible to antibodies than in BA.1. Binding and pseudovirus neutralization assays reveal extensive immune evasion while defining epitopes of two outer RBD face-binding antibodies, DH1044 and DH1193, that neutralize both BA.1 and BA.2. Taken together, our results indicate that stabilization of the closed state through interprotomer RBD-RBD packing is a hallmark of the Omicron variant and show differences in key functional regions in the BA.1 and BA.2 S proteins.
Organizational Affiliation:
Duke Human Vaccine Institute, Durham, NC 27710, USA.