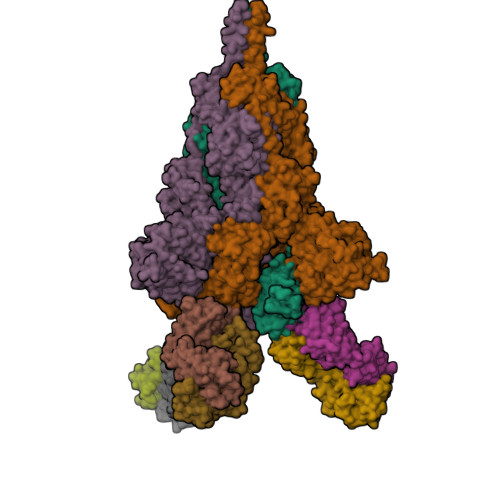
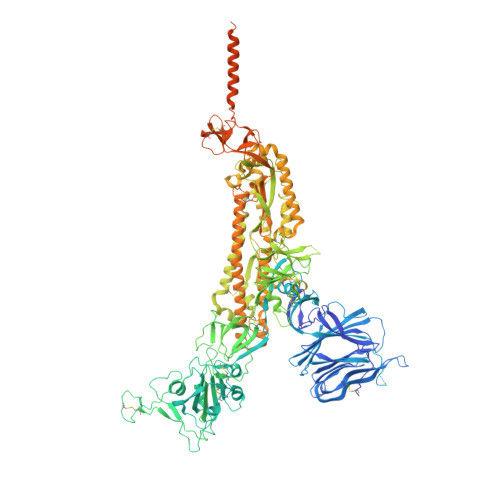
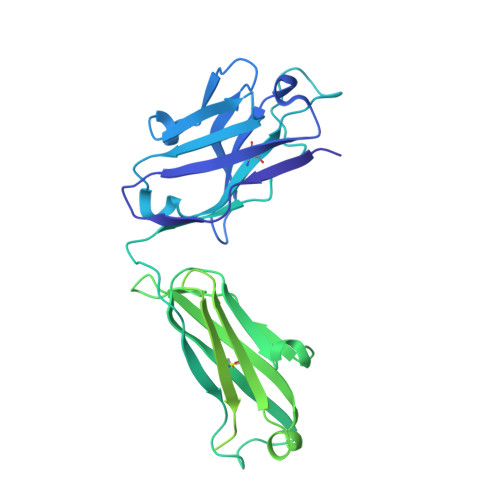
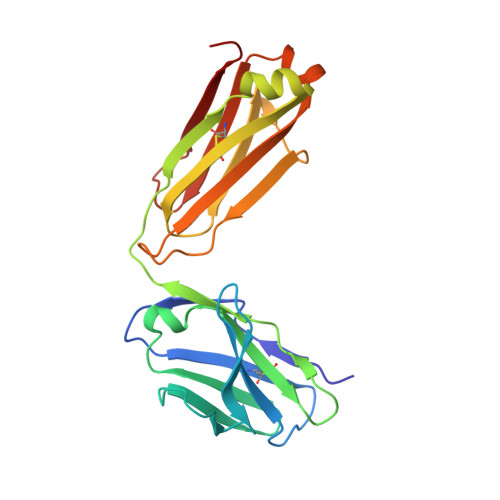
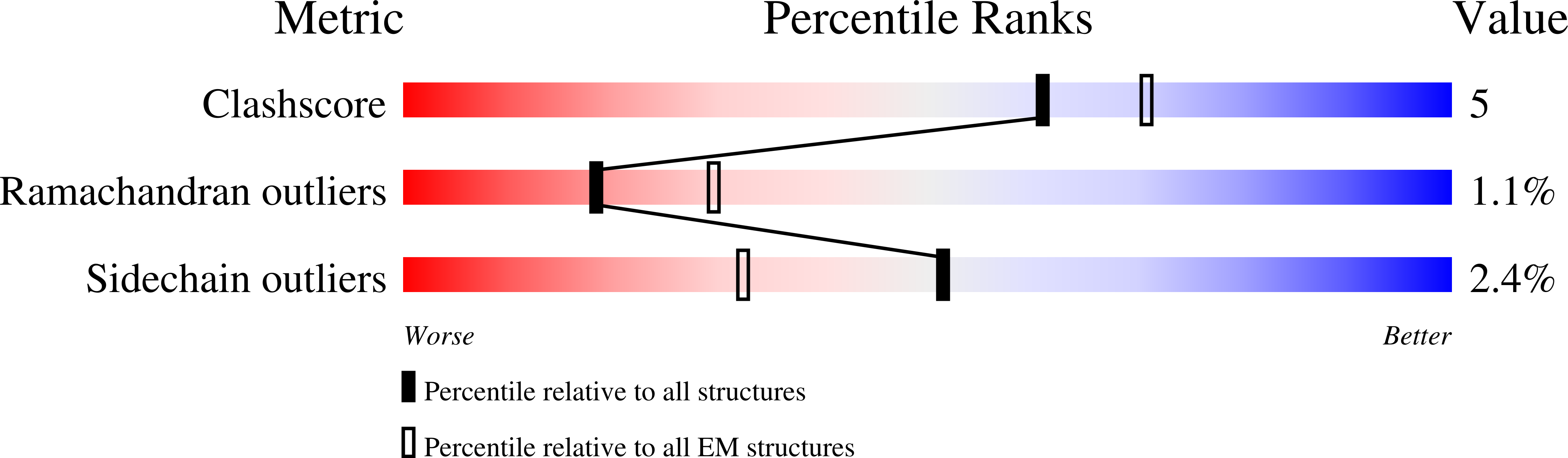An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Luo, S., Zhang, J., Kreutzberger, A.J.B., Eaton, A., Edwards, R.J., Jing, C., Dai, H.Q., Sempowski, G.D., Cronin, K., Parks, R., Ye, A.Y., Mansouri, K., Barr, M., Pishesha, N., Williams, A.C., Vieira Francisco, L., Saminathan, A., Peng, H., Batra, H., Bellusci, L., Khurana, S., Alam, S.M., Montefiori, D.C., Saunders, K.O., Tian, M., Ploegh, H., Kirchhausen, T., Chen, B., Haynes, B.F., Alt, F.W.(2022) Sci Immunol 7: eadd5446-eadd5446
- PubMed: 35951767
- DOI: https://doi.org/10.1126/sciimmunol.add5446
- Primary Citation of Related Structures:
7UPW, 7UPX, 7UPY - PubMed Abstract:
SARS-CoV-2 Omicron subvariants have generated a worldwide health crisis due to resistance to most approved SARS-CoV-2 neutralizing antibodies and evasion of vaccination-induced antibodies. To manage Omicron subvariants and prepare for new ones, additional means of isolating broad and potent humanized SARS-CoV-2 neutralizing antibodies are desirable. Here, we describe a mouse model in which the primary B cell receptor (BCR) repertoire is generated solely through V(D)J recombination of a human V H 1-2 heavy chain (HC) and, substantially, a human Vκ1-33 light chain (LC). Thus, primary humanized BCR repertoire diversity in these mice derives from immensely diverse HC and LC antigen-contact CDR3 sequences generated by nontemplated junctional modifications during V(D)J recombination. Immunizing this mouse model with SARS-CoV-2 (Wuhan-Hu-1) spike protein immunogens elicited several V H 1-2/Vκ1-33-based neutralizing antibodies that bound RBD in a different mode from each other and from those of many prior patient-derived V H 1-2-based neutralizing antibodies. Of these, SP1-77 potently and broadly neutralized all SARS-CoV-2 variants through BA.5. Cryo-EM studies revealed that SP1-77 bound RBD away from the receptor-binding motif via a CDR3-dominated recognition mode. Lattice light-sheet microscopy-based studies showed that SP1-77 did not block ACE2-mediated viral attachment or endocytosis but rather blocked viral-host membrane fusion. The broad and potent SP1-77 neutralization activity and nontraditional mechanism of action suggest that it might have therapeutic potential. Likewise, the SP1-77 binding epitope may inform vaccine strategies. Last, the type of humanized mouse models that we have described may contribute to identifying therapeutic antibodies against future SARS-CoV-2 variants and other pathogens.
Organizational Affiliation:
Howard Hughes Medical Institute, Program in Cellular and Molecular Medicine, Boston Children's Hospital, Boston, MA 02115, USA.