Structural basis of Streptomyces transcription activation by zinc uptake regulator.
Yang, X., Wang, Y., Liu, G., Deng, Z., Lin, S., Zheng, J.(2022) Nucleic Acids Res 50: 8363-8376
- PubMed: 35871291
- DOI: https://doi.org/10.1093/nar/gkac627
- Primary Citation of Related Structures:
7VO0, 7VO9, 7VPD, 7VPZ, 7X74, 7X75, 7X76 - PubMed Abstract:
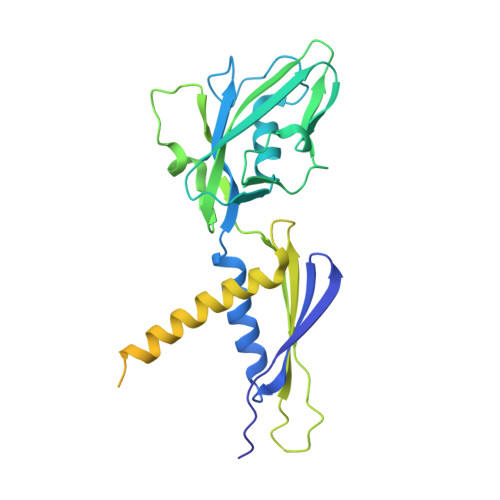
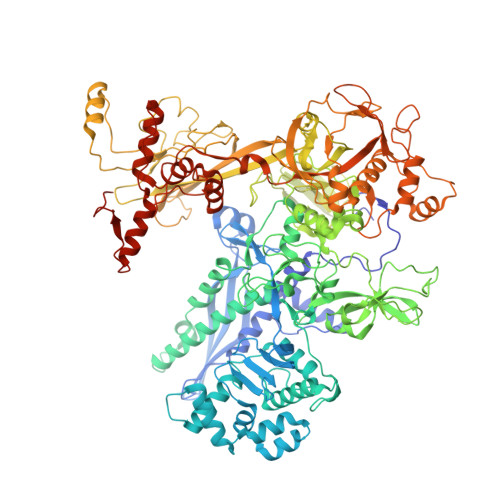
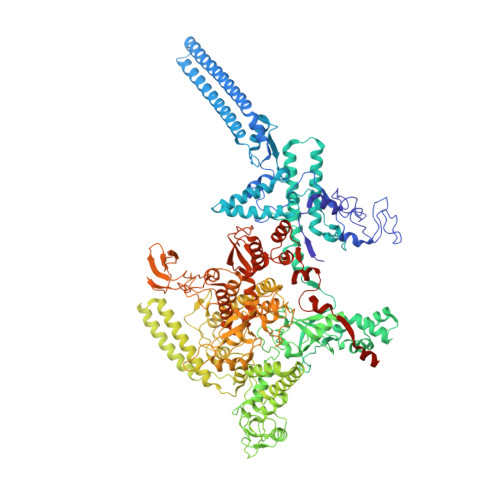
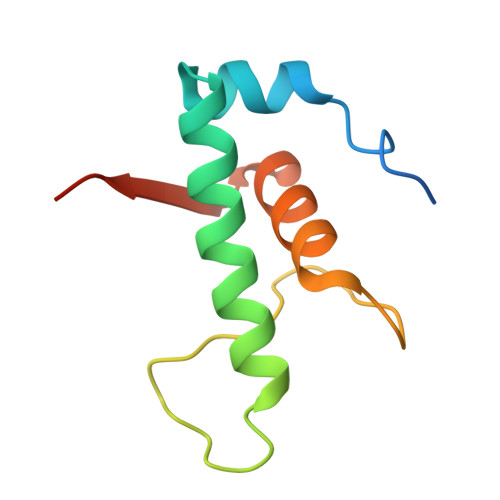
Streptomyces coelicolor (Sc) is a model organism of actinobacteria to study morphological differentiation and production of bioactive metabolites. Sc zinc uptake regulator (Zur) affects both processes by controlling zinc homeostasis. It activates transcription by binding to palindromic Zur-box sequences upstream of -35 elements. Here we deciphered the molecular mechanism by which ScZur interacts with promoter DNA and Sc RNA polymerase (RNAP) by cryo-EM structures and biochemical assays. The ScZur-DNA structures reveal a sequential and cooperative binding of three ScZur dimers surrounding a Zur-box spaced 8 nt upstream from a -35 element. The ScRNAPσHrdB-Zur-DNA structures define protein-protein and protein-DNA interactions involved in the principal housekeeping σHrdB-dependent transcription initiation from a noncanonical promoter with a -10 element lacking the critical adenine residue at position -11 and a TTGCCC -35 element deviating from the canonical TTGACA motif. ScZur interacts with the C-terminal domain of ScRNAP α subunit (αCTD) in a complex structure trapped in an active conformation. Key ScZur-αCTD interfacial residues accounting for ScZur-dependent transcription activation were confirmed by mutational studies. Together, our structural and biochemical results provide a comprehensive model for transcription activation of Zur family regulators.
Organizational Affiliation:
State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 200240, China.