SARS-CoV-2 hijacks neutralizing dimeric IgA for nasal infection and injury in Syrian hamsters 1 .
Zhou, B., Zhou, R., Chan, J.F., Zeng, J., Zhang, Q., Yuan, S., Liu, L., Robinot, R., Shan, S., Liu, N., Ge, J., Kwong, H.Y., Zhou, D., Xu, H., Chan, C.C., Poon, V.K., Chu, H., Yue, M., Kwan, K.Y., Chan, C.Y., Chan, C.C., Chik, K.K., Du, Z., Au, K.K., Huang, H., Man, H.O., Cao, J., Li, C., Wang, Z., Zhou, J., Song, Y., Yeung, M.L., To, K.K., Ho, D.D., Chakrabarti, L.A., Wang, X., Zhang, L., Yuen, K.Y., Chen, Z.(2023) Emerg Microbes Infect 12: 2245921-2245921
- PubMed: 37542391
- DOI: https://doi.org/10.1080/22221751.2023.2245921
- Primary Citation of Related Structures:
7WH8, 7WHB, 7WHD - PubMed Abstract:
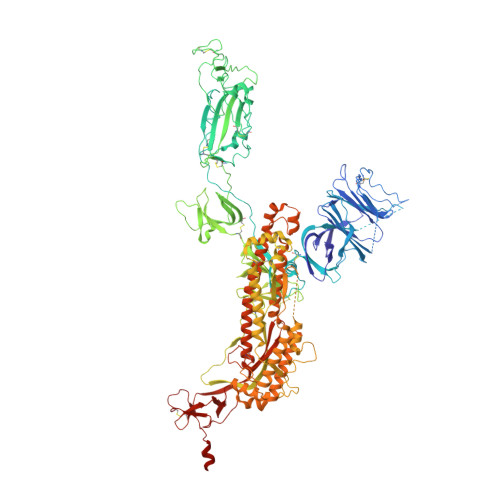
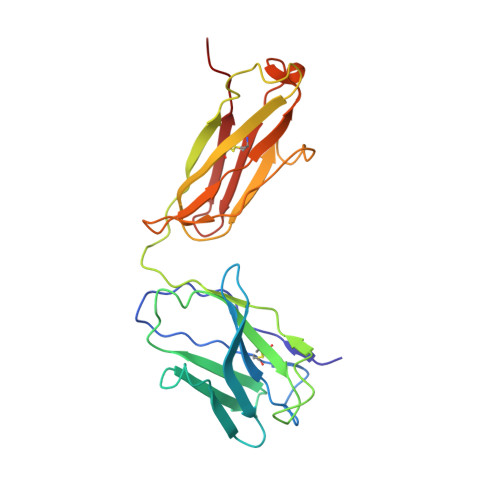
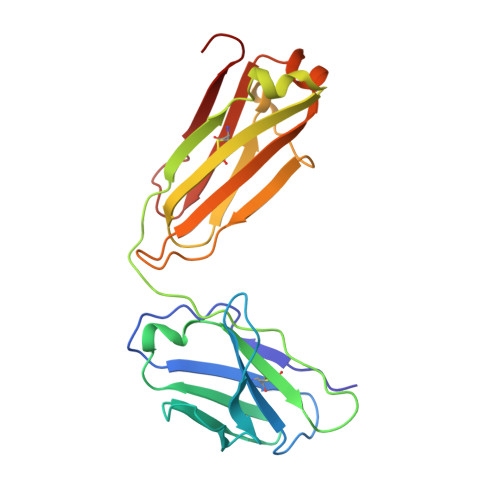
Prevention of robust severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection in nasal turbinate (NT) requires in vivo evaluation of IgA neutralizing antibodies. Here, we report the efficacy of receptor binding domain (RBD)-specific monomeric B8-mIgA1 and B8-mIgA2, and dimeric B8-dIgA1, B8-dIgA2 and TH335-dIgA1 against intranasal SARS-CoV-2 challenge in Syrian hamsters. These antibodies exhibited comparable neutralization potency against authentic virus by competing with human angiotensin converting enzyme-2 (ACE2) receptor for RBD binding. While reducing viral loads in lungs significantly, prophylactic intranasal B8-dIgA unexpectedly led to high amount of infectious viruses and extended damage in NT compared to controls. Mechanistically, B8-dIgA failed to inhibit SARS-CoV-2 cell-to-cell transmission, but was hijacked by the virus through dendritic cell-mediated trans-infection of NT epithelia leading to robust nasal infection. Cryo-EM further revealed B8 as a class II antibody binding trimeric RBDs in 3-up or 2-up/1-down conformation. Neutralizing dIgA, therefore, may engage an unexpected mode of SARS-CoV-2 nasal infection and injury.
Organizational Affiliation:
AIDS Institute, School of Clinical Medicine, Li Ka Shing Faculty of Medicine, the University of Hong Kong, Pokfulam, Hong Kong Special Administrative Region (SAR), People's Republic of China.