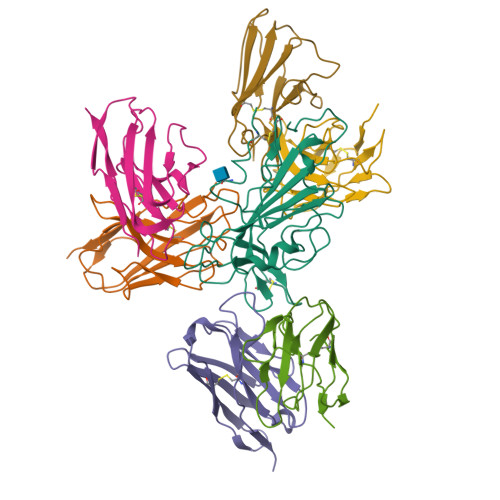
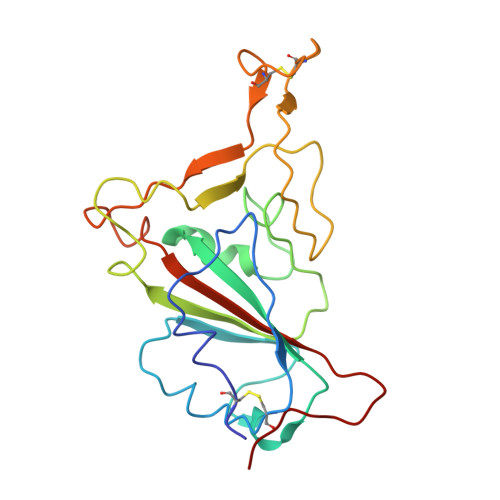
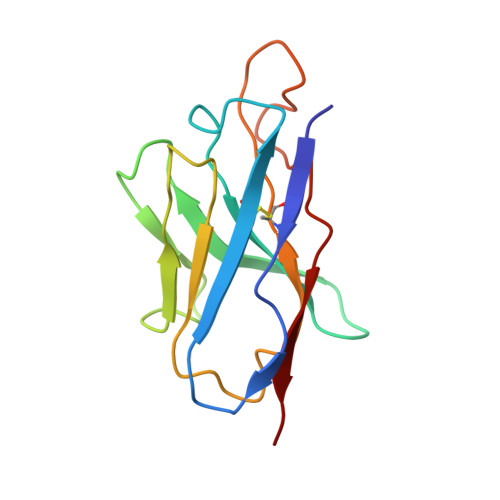
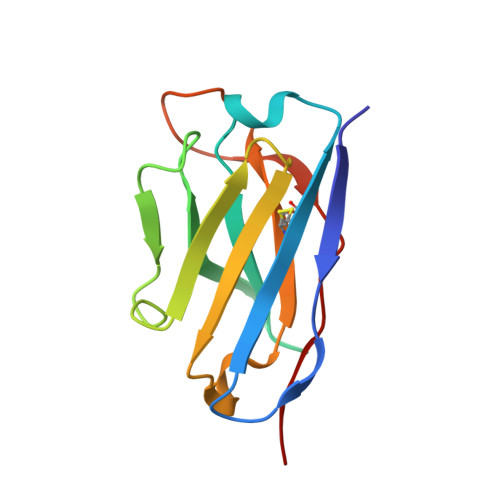
Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Wang, S., Sun, H., Zhang, Y., Yuan, L., Wang, Y., Zhang, T., Wang, S., Zhang, J., Yu, H., Xiong, H., Tang, Z., Liu, L., Huang, Y., Chen, X., Li, T., Ying, D., Liu, C., Chen, Z., Yuan, Q., Zhang, J., Cheng, T., Li, S., Guan, Y., Zheng, Q., Zheng, Z., Xia, N.(2022) Cell Rep 39: 110862-110862
- PubMed: 35594869
- DOI: https://doi.org/10.1016/j.celrep.2022.110862
- Primary Citation of Related Structures:
7WHZ, 7WI0 - PubMed Abstract:
The rapidly spreading Omicron variant is highly resistant to vaccines, convalescent sera, and neutralizing antibodies (nAbs), highlighting the urgent need for potent therapeutic nAbs. Here, a panel of human nAbs from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) convalescent patients show diverse neutralization against Omicron, of which XMA01 and XMA04 maintain nanomolar affinities and excellent neutralization (half maximal inhibitory concentration [IC50]: ∼20 ng/mL). nAb XMA09 shows weak but unattenuated neutralization against all variants of concern (VOCs) as well as SARS-CoV. Structural analysis reveals that the above three antibodies could synergistically bind to the receptor-binding domains (RBDs) of both wild-type and Omicron spikes and defines the critical determinants for nAb-mediated broad neutralizations. Three nAbs confer synergistic neutralization against Omicron, resulting from the inter-antibody interaction between XMA04 and XMA01(or XMA09). Furthermore, the XMA01/XMA04 cocktail provides synergistic protection against Beta and Omicron variant infections in hamsters. In summary, our results provide insights for the rational design of antibody cocktail therapeutics or universal vaccines against Omicron.
Organizational Affiliation:
State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics, Xiamen 361102, China; National Institute of Diagnostics and Vaccine Development in Infectious Diseases, School of Public Health, School of Life Sciences, Xiamen University, Xiamen 361102, China.