Structural basis for the regulation mechanism of a hyper polarization-activated K+ channel AKT1 by AtKC1
Dongliang, L., Zijie, Z., Yannan, Q., Yuyue, T., Huaizong, S.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Potassium channel AKT1 | 885 | Arabidopsis thaliana | Mutation(s): 0 Gene Names: AKT1, At2g26650, F18A8.2 Membrane Entity: Yes | 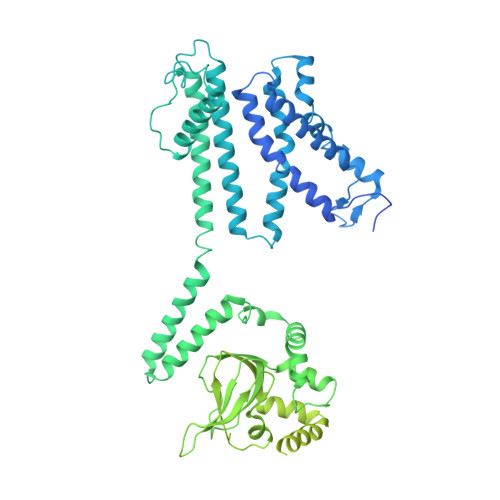 | |
UniProt | |||||
Find proteins for Q38998 (Arabidopsis thaliana) Explore Q38998 Go to UniProtKB: Q38998 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q38998 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Potassium channel KAT3 | 690 | Arabidopsis thaliana | Mutation(s): 0 Gene Names: KAT3, AKT4, KC1, At4g32650, F4D11.150 Membrane Entity: Yes | 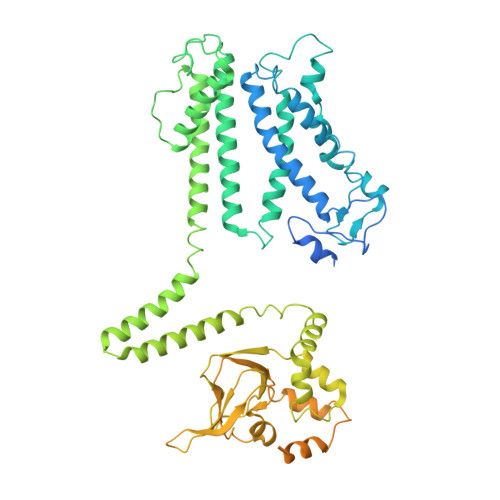 | |
UniProt | |||||
Find proteins for P92960 (Arabidopsis thaliana) Explore P92960 Go to UniProtKB: P92960 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P92960 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| POV Query on POV | E [auth A], I [auth B], J [auth C], K [auth D] | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate C42 H82 N O8 P WTJKGGKOPKCXLL-PFDVCBLKSA-N |  | ||
| K Query on K | F [auth A], G [auth A], H [auth A] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | -- |