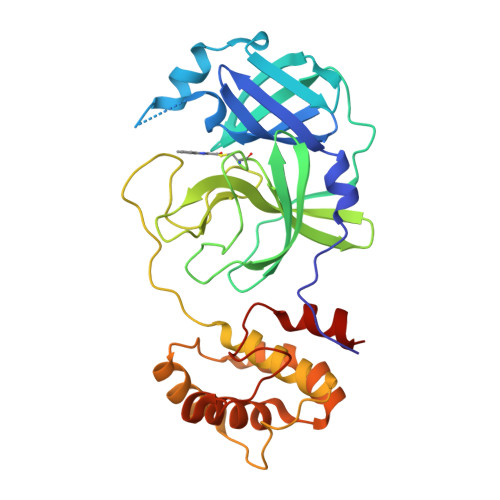
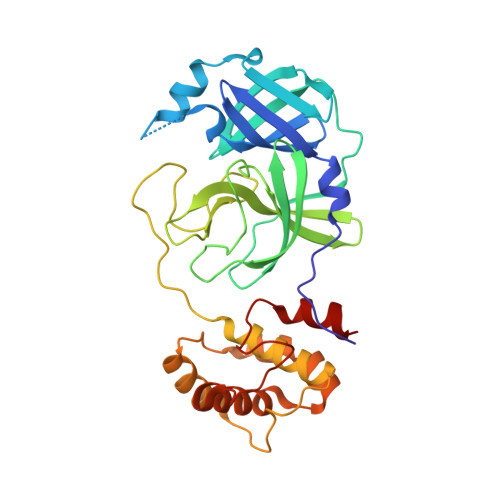
Small-Molecule Thioesters as SARS-CoV-2 Main Protease Inhibitors: Enzyme Inhibition, Structure-Activity Relationships, Antiviral Activity, and X-ray Structure Determination.
Pillaiyar, T., Flury, P., Kruger, N., Su, H., Schakel, L., Barbosa Da Silva, E., Eppler, O., Kronenberger, T., Nie, T., Luedtke, S., Rocha, C., Sylvester, K., Petry, M.R.I., McKerrow, J.H., Poso, A., Pohlmann, S., Gutschow, M., O'Donoghue, A.J., Xu, Y., Muller, C.E., Laufer, S.A.(2022) J Med Chem 65: 9376-9395
- PubMed: 35709506
- DOI: https://doi.org/10.1021/acs.jmedchem.2c00636
- Primary Citation of Related Structures:
7X6J, 7X6K - PubMed Abstract:
The main protease (M pro , 3CL pro ) of SARS-CoV-2 is an attractive target in coronaviruses because of its crucial involvement in viral replication and transcription. Here, we report on the design, synthesis, and structure-activity relationships of novel small-molecule thioesters as SARS-CoV-2 M pro inhibitors. Compounds 3w and 3x exhibited excellent SARS-CoV-2 M pro inhibition with k inac / K i of 58,700 M -1 s -1 ( K i = 0.0141 μM) and 27,200 M -1 s -1 ( K i = 0.0332 μM), respectively. In Calu-3 and Vero76 cells, compounds 3h , 3i, 3l , 3r , 3v , 3w , and 3x displayed antiviral activity in the nanomolar range without host cell toxicity. Co-crystallization of 3w and 3af with SARS-CoV-2 M pro was accomplished, and the X-ray structures showed covalent binding with the catalytic Cys145 residue of the protease. The potent SARS-CoV-2 Mpro inhibitors also inhibited the M pro of other beta-coronaviruses, including SARS-CoV-1 and MERS-CoV, indicating that they might be useful to treat a broader range of coronaviral infections.
Organizational Affiliation:
Institute of Pharmacy, Pharmaceutical/Medicinal Chemistry and Tübingen Center for Academic Drug Discovery, Eberhard Karls University Tübingen, Auf der Morgenstelle 8, 72076 Tübingen, Germany. Cluster of Excellence iFIT (EXC 2180) "Image-Guided & Functionally Instructed Tumor Therapies", University of Tübingen, Tübingen 72076, Germany.