Engineering an autonomous VH domain to modulate intracellular pathways and to interrogate the eIF4F complex.
Frosi, Y., Lin, Y.C., Shimin, J., Ramlan, S.R., Hew, K., Engman, A.H., Pillai, A., Yeung, K., Cheng, Y.X., Cornvik, T., Nordlund, P., Goh, M., Lama, D., Gates, Z.P., Verma, C.S., Thean, D., Lane, D.P., Asial, I., Brown, C.J.(2022) Nat Commun 13: 4854-4854
- PubMed: 35982046
- DOI: https://doi.org/10.1038/s41467-022-32463-1
- Primary Citation of Related Structures:
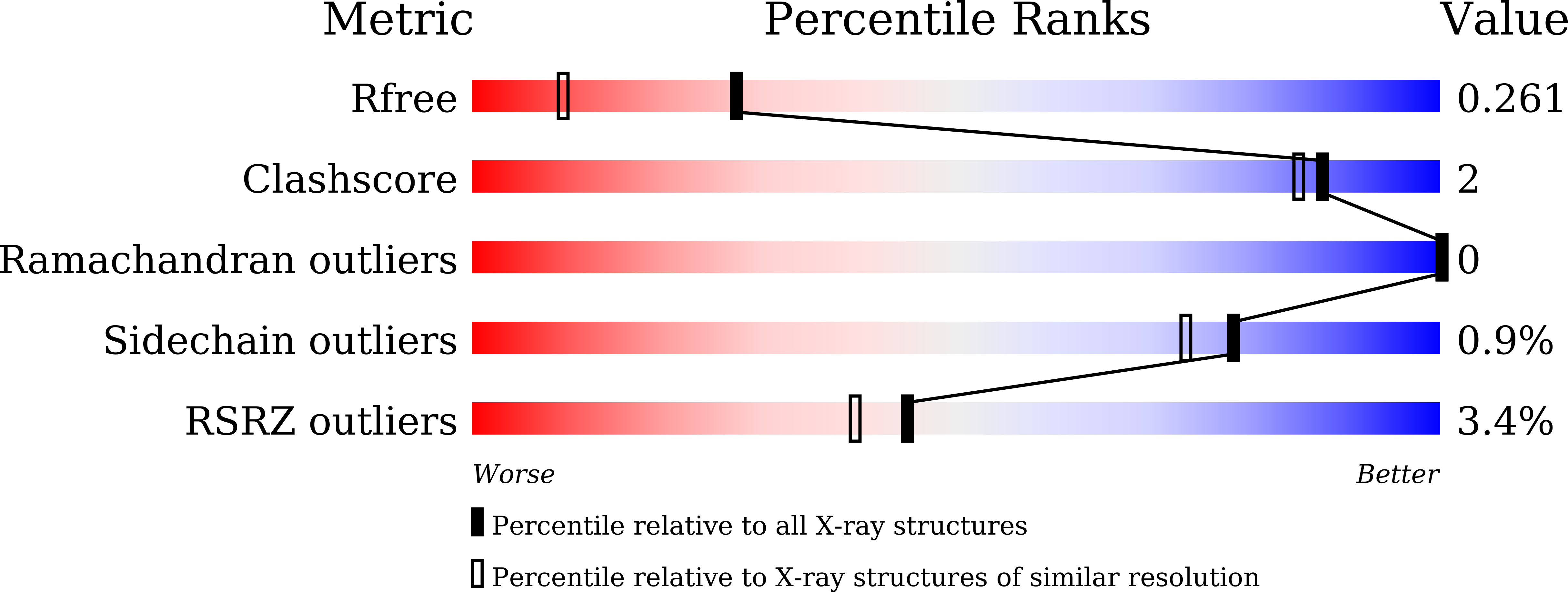
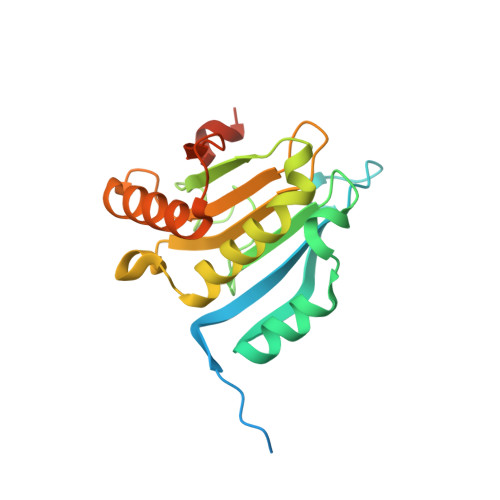
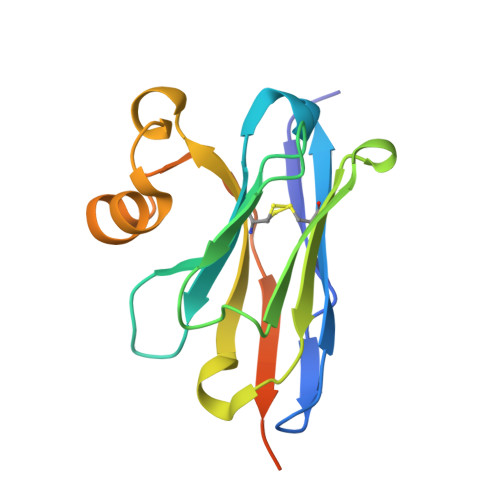
7D8B, 7XTP - PubMed Abstract:
An attractive approach to target intracellular macromolecular interfaces and to model putative drug interactions is to design small high-affinity proteins. Variable domains of the immunoglobulin heavy chain (VH domains) are ideal miniproteins, but their development has been restricted by poor intracellular stability and expression. Here we show that an autonomous and disufhide-free VH domain is suitable for intracellular studies and use it to construct a high-diversity phage display library. Using this library and affinity maturation techniques we identify VH domains with picomolar affinity against eIF4E, a protein commonly hyper-activated in cancer. We demonstrate that these molecules interact with eIF4E at the eIF4G binding site via a distinct structural pose. Intracellular overexpression of these miniproteins reduce cellular proliferation and expression of malignancy-related proteins in cancer cell lines. The linkage of high-diversity in vitro libraries with an intracellularly expressible miniprotein scaffold will facilitate the discovery of VH domains suitable for intracellular applications.
Organizational Affiliation:
p53 Laboratory (A*STAR), 8A Biomedical Grove, #06-04/05, Neuros/Immunos, 138648, Singapore.