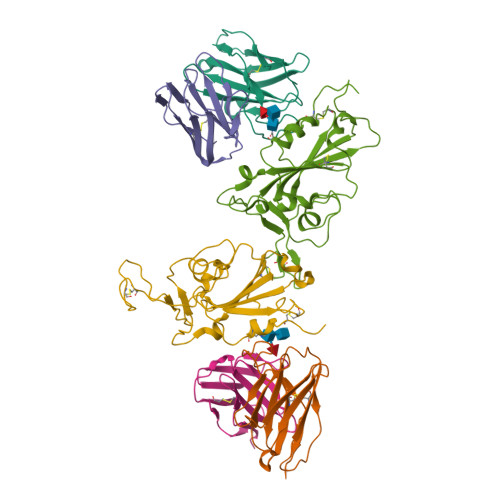
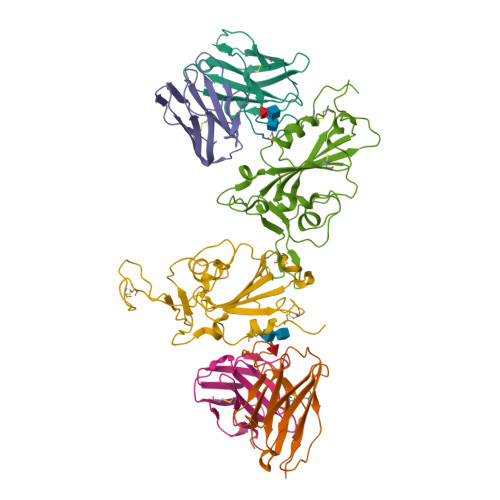
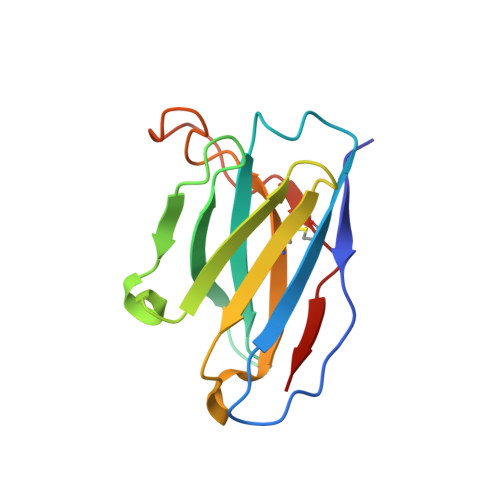
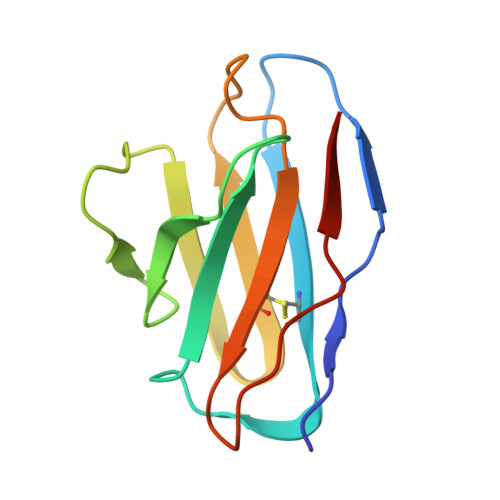
Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Zhao, Z., Zhou, J., Tian, M., Huang, M., Liu, S., Xie, Y., Han, P., Bai, C., Han, P., Zheng, A., Fu, L., Gao, Y., Peng, Q., Li, Y., Chai, Y., Zhang, Z., Zhao, X., Song, H., Qi, J., Wang, Q., Wang, P., Gao, G.F.(2022) Nat Commun 13: 4958-4958
- PubMed: 36002453
- DOI: https://doi.org/10.1038/s41467-022-32665-7
- Primary Citation of Related Structures:
7Y9S, 7Y9Z, 7YA0, 7YA1, 7YAD - PubMed Abstract:
Omicron SARS-CoV-2 is rapidly spreading worldwide. To delineate the impact of emerging mutations on spike's properties, we performed systematic structural analyses on apo Omicron spike and its complexes with human ACE2 or S309 neutralizing antibody (NAb) by cryo-EM. The Omicron spike preferentially adopts the one-RBD-up conformation both before and after ACE2 binding, which is in sharp contrast to the orchestrated conformational changes to create more up-RBDs upon ACE2 binding as observed in the prototype and other four variants of concern (VOCs). Furthermore, we found that S371L, S373P and S375F substitutions enhance the stability of the one-RBD-up conformation to prevent exposing more up-RBDs triggered by ACE2 binding. The increased stability of the one-RBD-up conformation restricts the accessibility of S304 NAb, which targets a cryptic epitope in the closed conformation, thus facilitating the immune evasion by Omicron. These results expand our understanding of Omicron spike's conformation, receptor binding and antibody evasion mechanism.
Organizational Affiliation:
CAS Key Laboratory of Pathogen Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, 100101, China.