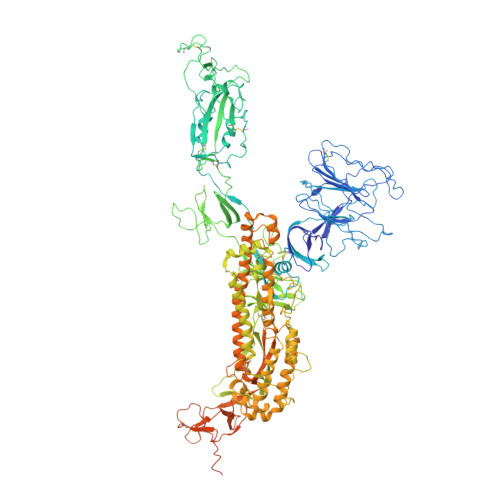
Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cao, Y., Song, W., Wang, L., Liu, P., Yue, C., Jian, F., Yu, Y., Yisimayi, A., Wang, P., Wang, Y., Zhu, Q., Deng, J., Fu, W., Yu, L., Zhang, N., Wang, J., Xiao, T., An, R., Wang, J., Liu, L., Yang, S., Niu, X., Gu, Q., Shao, F., Hao, X., Meng, B., Gupta, R.K., Jin, R., Wang, Y., Xie, X.S., Wang, X.(2022) Cell Host Microbe 30: 1527
- PubMed: 36270286
- DOI: https://doi.org/10.1016/j.chom.2022.09.018
- Primary Citation of Related Structures:
7YQT, 7YQU, 7YQV, 7YQW, 7YQX, 7YQY, 7YQZ, 7YR0, 7YR1, 7YR2, 7YR3 - PubMed Abstract:
Recently emerged SARS-CoV-2 Omicron subvariant, BA.2.75, displayed a growth advantage over circulating BA.2.38, BA.2.76, and BA.5 in India. However, the underlying mechanisms for enhanced infectivity, especially compared with BA.5, remain unclear. Here, we show that BA.2.75 exhibits substantially higher affinity for host receptor angiotensin-converting enzyme 2 (ACE2) than BA.5 and other variants. Structural analyses of BA.2.75 spike shows its decreased thermostability and increased frequency of the receptor binding domain (RBD) in the "up" conformation under acidic conditions, suggesting enhanced low-pH-endosomal cell entry. Relative to BA.4/BA.5, BA.2.75 exhibits reduced evasion of humoral immunity from BA.1/BA.2 breakthrough-infection convalescent plasma but greater evasion of Delta breakthrough-infection convalescent plasma. BA.5 breakthrough-infection plasma also exhibits weaker neutralization against BA.2.75 than BA.5, mainly due to BA.2.75's distinct neutralizing antibody (NAb) escape pattern. Antibody therapeutics Evusheld and Bebtelovimab remain effective against BA.2.75. These results suggest BA.2.75 may prevail after BA.4/BA.5, and its increased receptor-binding capability could support further immune-evasive mutations.
Organizational Affiliation:
Biomedical Pioneering Innovation Center (BIOPIC), Peking University, Beijing 100871, China; Changping Laboratory, Beijing 102206, China. Electronic address: yunlongcao@pku.edu.cn.