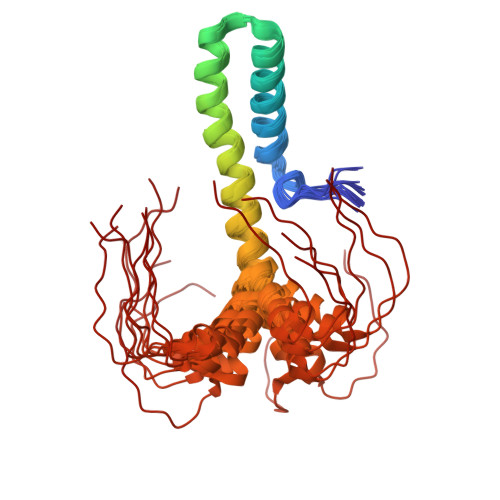
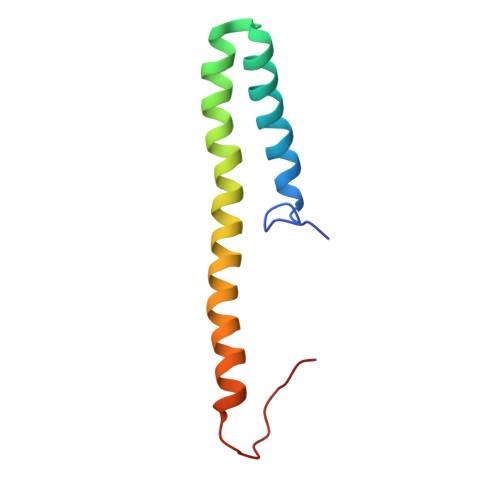
SARS-CoV-2 Nsp8 N-terminal domain folds autonomously and binds dsRNA.
Trevino, M.A., Pantoja-Uceda, D., Laurents, D.V., Mompean, M.(2023) Nucleic Acids Res 51: 10041-10048
- PubMed: 37665006
- DOI: https://doi.org/10.1093/nar/gkad714
- Primary Citation of Related Structures:
7YWR - PubMed Abstract:
The SARS-CoV-2 Nsp8 protein is a critical component of the RNA replicase, as its N-terminal domain (NTD) anchors Nsp12, the RNA, and Nsp13. Whereas its C-terminal domain (CTD) structure is well resolved, there is an open debate regarding the conformation adopted by the NTD as it is predicted as disordered but found in a variety of complex-dependent conformations or missing from many other structures. Using NMR spectroscopy, we show that the SARS CoV-2 Nsp8 NTD features both well folded secondary structure and disordered segments. Our results suggest that while part of this domain corresponding to two long α-helices forms autonomously, the folding of other segments would require interaction with other replicase components. When isolated, the α-helix population progressively declines towards the C-termini but surprisingly binds dsRNA while preserving structural disorder.
Organizational Affiliation:
"Blas Cabrera" Institute for Physical Chemistry, Spanish National Research Council, Serrano 119, Madrid 28006, Spain.