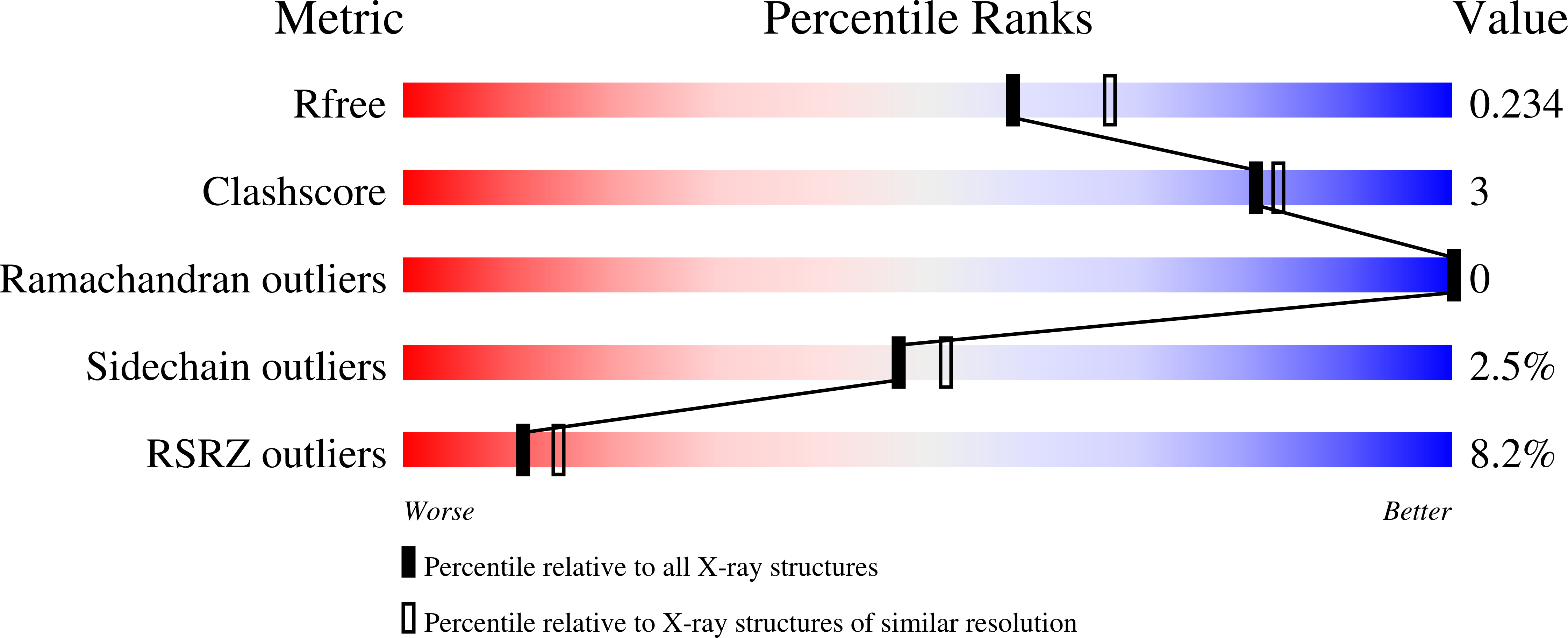
Structural and Biochemical Investigation of the Heterodimeric Murine tRNA-Guanine Transglycosylase.
Sebastiani, M., Behrens, C., Dorr, S., Gerber, H.D., Benazza, R., Hernandez-Alba, O., Cianferani, S., Klebe, G., Heine, A., Reuter, K.(2022) ACS Chem Biol 17: 2229-2247
- PubMed: 35815944
- DOI: https://doi.org/10.1021/acschembio.2c00368
- Primary Citation of Related Structures:
6H62, 7B2I, 7OV9, 7OVO, 7OVS, 7OWZ - PubMed Abstract:
In tRNA Asp , tRNA Asn , tRNA Tyr , and tRNA His of most bacteria and eukaryotes, the anticodon wobble position may be occupied by the modified nucleoside queuosine, which affects the speed and the accuracy of translation. Since eukaryotes are not able to synthesize queuosine de novo, they have to salvage queuine (the queuosine base) as a micronutrient from food and/or the gut microbiome. The heterodimeric Zn 2+ containing enzyme tRNA-guanine transglycosylase (TGT) catalyzes the insertion of queuine into the above-named tRNAs in exchange for the genetically encoded guanine. This enzyme has attracted medical interest since it was shown to be potentially useful for the treatment of multiple sclerosis. In addition, TGT inactivation via gene knockout leads to the suppressed cell proliferation and migration of certain breast cancer cells, which may render this enzyme a potential target for the design of compounds supporting breast cancer therapy. As a prerequisite to fully exploit the medical potential of eukaryotic TGT, we have determined and analyzed a number of crystal structures of the functional murine TGT with and without bound queuine. In addition, we have investigated the importance of two residues of its non-catalytic subunit on dimer stability and determined the Michaelis-Menten parameters of murine TGT with respect to tRNA and several natural and artificial nucleobase substrates. Ultimately, on the basis of available TGT crystal structures, we provide an entirely conclusive reaction mechanism for this enzyme, which in detail explains why the TGT-catalyzed insertion of some nucleobases into tRNA occurs reversibly while that of others is irreversible.
Organizational Affiliation:
Institut für Pharmazeutische Chemie, Philipps-Universität Marburg, Marbacher Weg 8, D-35037 Marburg, Germany.