Human 14-3-3 Proteins Site-selectively Bind the Mutational Hotspot Region of SARS-CoV-2 Nucleoprotein Modulating its Phosphoregulation.
Tugaeva, K.V., Sysoev, A.A., Kapitonova, A.A., Smith, J.L.R., Zhu, P., Cooley, R.B., Antson, A.A., Sluchanko, N.N.(2023) J Mol Biol 435: 167891-167891
- PubMed: 36427566
- DOI: https://doi.org/10.1016/j.jmb.2022.167891
- Primary Citation of Related Structures:
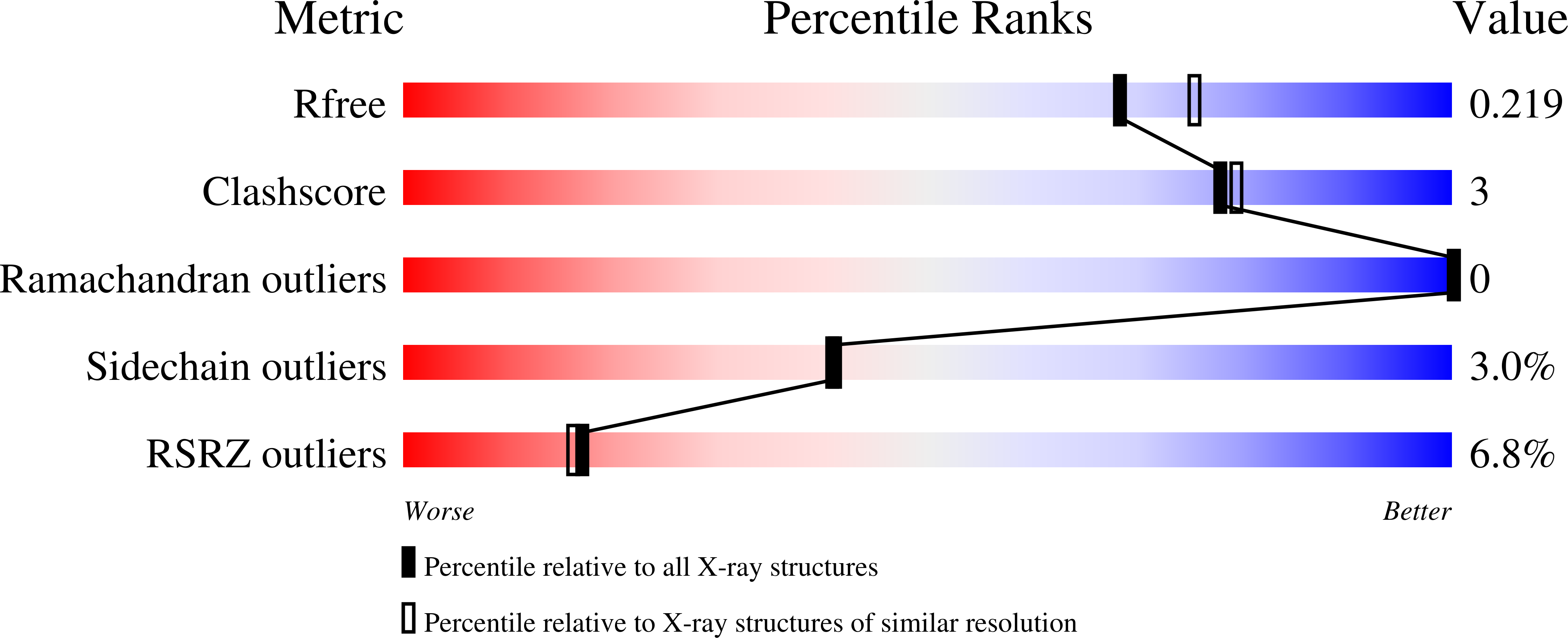
7QIK, 7QIP - PubMed Abstract:
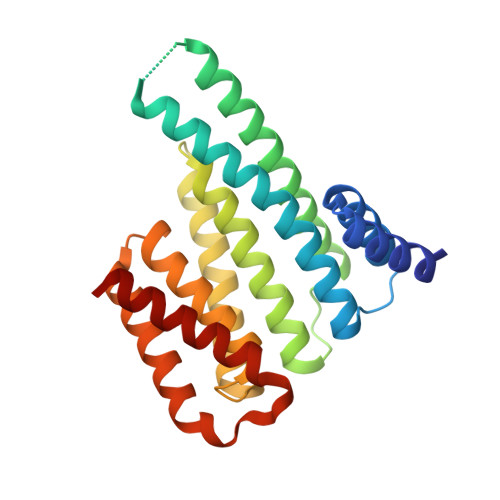
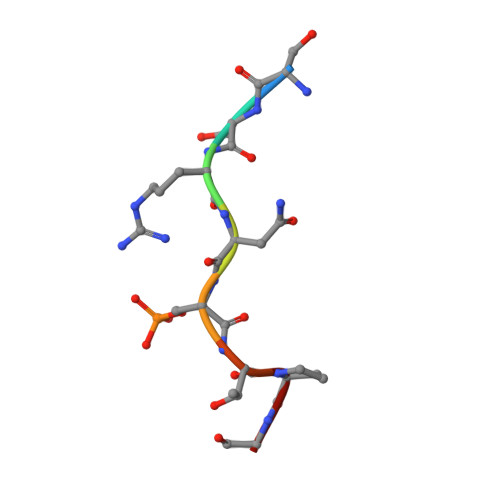
Phosphorylation of SARS-CoV-2 nucleoprotein recruits human cytosolic 14-3-3 proteins playing a well-recognized role in replication of many viruses. Here we use genetic code expansion to demonstrate that 14-3-3 binding is triggered by phosphorylation of SARS-CoV-2 nucleoprotein at either of two pseudo-repeats centered at Ser197 and Thr205. According to fluorescence anisotropy measurements, the pT205-motif,presentin SARS-CoV-2 but not in SARS-CoV, is preferred over the pS197-motif by all seven human 14-3-3 isoforms, which collectively display an unforeseen pT205/pS197 peptide binding selectivity hierarchy. Crystal structures demonstrate that pS197 and pT205 are mutually exclusive 14-3-3-binding sites, whereas SAXS and biochemical data obtained on the full protein-protein complex indicate that 14-3-3 binding occludes the Ser/Arg-rich region of the nucleoprotein, inhibiting its dephosphorylation. This Ser/Arg-rich region is highly prone to mutations, as exemplified by the Omicron and Delta variants, with our data suggesting that the strength of 14-3-3/nucleoprotein interaction can be linked with the replicative fitness of the virus.
Organizational Affiliation:
A.N. Bach Institute of Biochemistry, Federal Research Center of Biotechnology of the Russian Academy of Sciences, 119071 Moscow, Russia.