Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
Scaletti, E.R., Pettersson, P., Patrick, J., Shilling, P.J., Westergren, R.G., Daley, D.O., Maler, L., Widmalm, G., Stenmark, P.(2023) J Biological Chem 299: 105256-105256
- PubMed: 37716703
- DOI: https://doi.org/10.1016/j.jbc.2023.105256
- Primary Citation of Related Structures:
8B5Q, 8B5S, 8B62, 8B63 - PubMed Abstract:
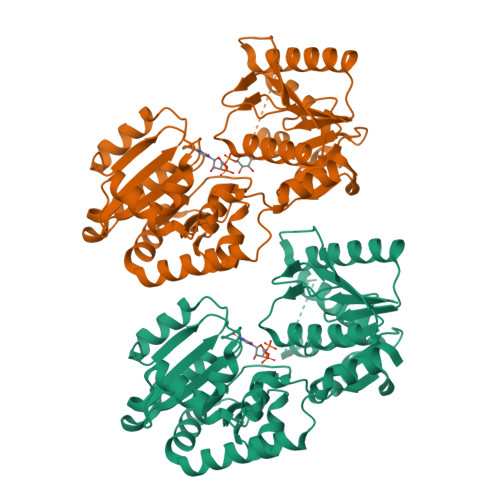
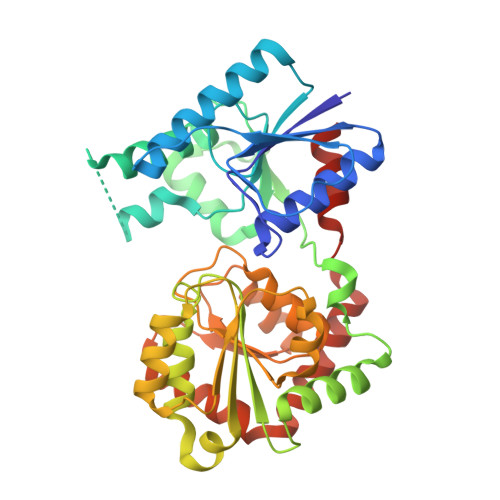
The glycosyltransferase WaaG in Pseudomonas aeruginosa (PaWaaG) is involved in the synthesis of the core region of lipopolysaccharides. It is a promising target for developing adjuvants that could help in the uptake of antibiotics. Herein, we have determined structures of PaWaaG in complex with the nucleotide-sugars UDP-glucose, UDP-galactose, and UDP-GalNAc. Structural comparison with the homolog from Escherichia coli (EcWaaG) revealed five key differences in the sugar-binding pocket. Solution-state NMR analysis showed that WT PaWaaG specifically hydrolyzes UDP-GalNAc and unlike EcWaaG, does not hydrolyze UDP-glucose. Furthermore, we found that a PaWaaG mutant (Y97F/T208R/N282A/T283A/T285I) designed to resemble the EcWaaG sugar binding site, only hydrolyzed UDP-glucose, underscoring the importance of the identified amino acids in substrate specificity. However, neither WT PaWaaG nor the PaWaaG mutant capable of hydrolyzing UDP-glucose was able to complement an E. coli ΔwaaG strain, indicating that more remains to be uncovered about the function of PaWaaG in vivo. This structural and biochemical information will guide future structure-based drug design efforts targeting PaWaaG.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Stockholm University, Stockholm, Sweden.