Sulfonamidoboronic Acids as "Cross-Class" Inhibitors of an Expanded-Spectrum Class C Cephalosporinase, ADC-33, and a Class D Carbapenemase, OXA-24/40: Strategic Compound Design to Combat Resistance in Acinetobacter baumannii .
Introvigne, M.L., Beardsley, T.J., Fernando, M.C., Leonard, D.A., Wallar, B.J., Rudin, S.D., Taracila, M.A., Rather, P.N., Colquhoun, J.M., Song, S., Fini, F., Hujer, K.M., Hujer, A.M., Prati, F., Powers, R.A., Bonomo, R.A., Caselli, E.(2023) Antibiotics (Basel) 12
- PubMed: 37107006
- DOI: https://doi.org/10.3390/antibiotics12040644
- Primary Citation of Related Structures:
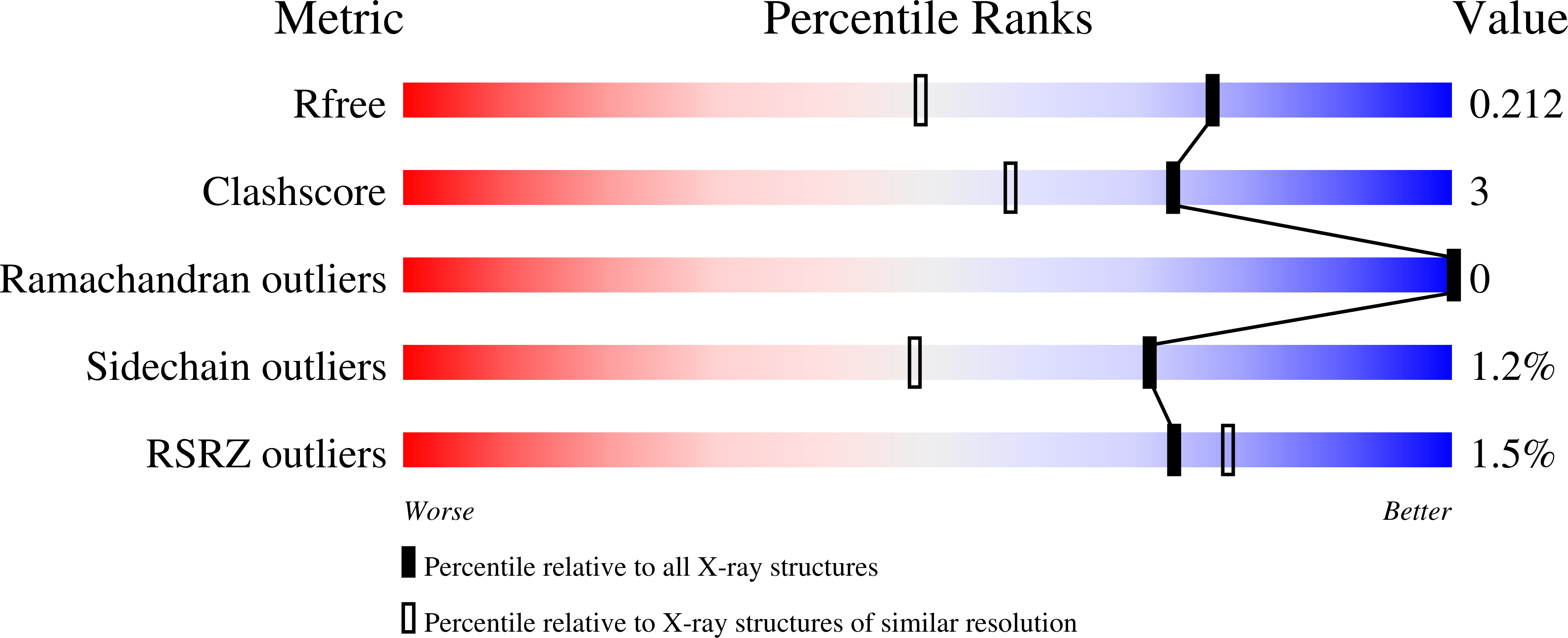
8CUL, 8CUM, 8CUO, 8CUP, 8CUQ - PubMed Abstract:
Acinetobacter baumannii is a Gram-negative organism listed as an urgent threat pathogen by the World Health Organization (WHO). Carbapenem-resistant A. baumannii (CRAB), especially, present therapeutic challenges due to complex mechanisms of resistance to β -lactams. One of the most important mechanisms is the production of β -lactamase enzymes capable of hydrolyzing β -lactam antibiotics. Co-expression of multiple classes of β -lactamases is present in CRAB; therefore, the design and synthesis of "cross-class" inhibitors is an important strategy to preserve the efficacy of currently available antibiotics. To identify new, nonclassical β -lactamase inhibitors, we previously identified a sulfonamidomethaneboronic acid CR167 active against Acinetobacter -derived class C β -lactamases (ADC-7). The compound demonstrated affinity for ADC-7 with a K i = 160 nM and proved to be able to decrease MIC values of ceftazidime and cefotaxime in different bacterial strains. Herein, we describe the activity of CR167 against other β -lactamases in A. baumannii : the cefepime-hydrolysing class C extended-spectrum β -lactamase (ESAC) ADC-33 and the carbapenem-hydrolyzing OXA-24/40 (class D). These investigations demonstrate CR167 as a valuable cross-class (C and D) inhibitor, and the paper describes our attempts to further improve its activity. Five chiral analogues of CR167 were rationally designed and synthesized. The structures of OXA-24/40 and ADC-33 in complex with CR167 and select chiral analogues were obtained. The structure activity relationships (SARs) are highlighted, offering insights into the main determinants for cross-class C/D inhibitors and impetus for novel drug design.
Organizational Affiliation:
Department of Life Sciences, Università di Modena e Reggio Emilia, Via Campi 103, 41125 Modena, Italy.