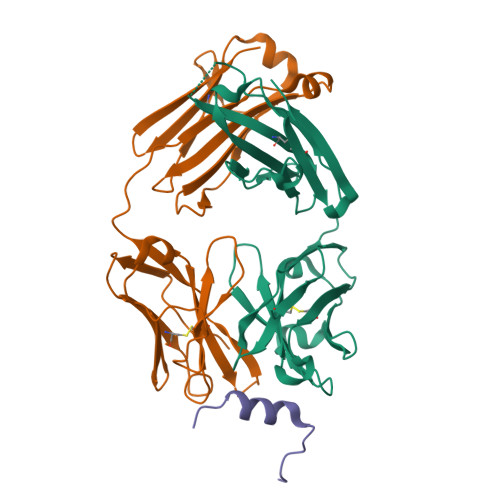
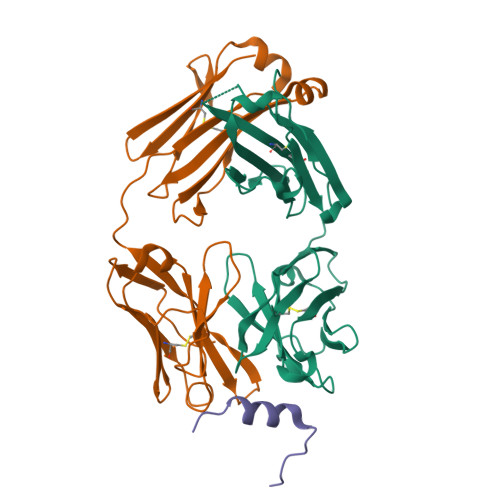
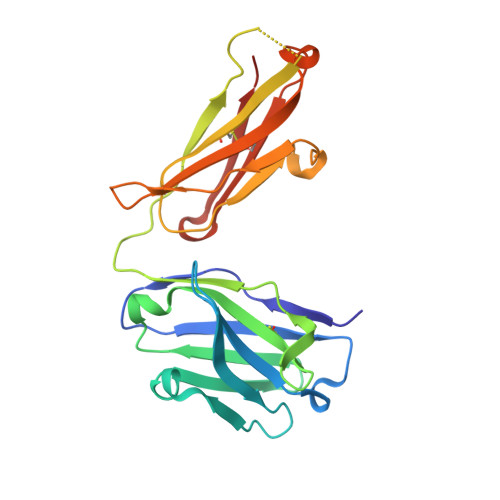
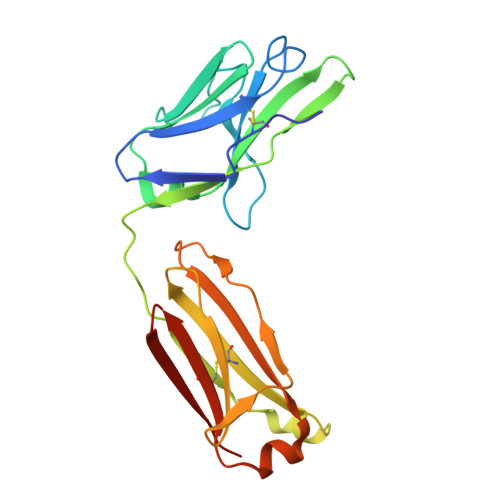
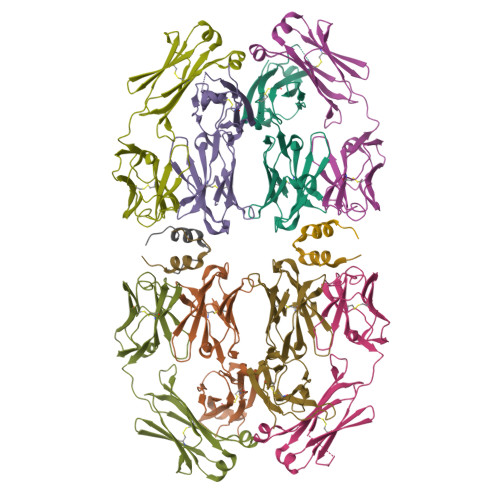Broadly neutralizing anti-S2 antibodies protect against all three human betacoronaviruses that cause deadly disease.
Zhou, P., Song, G., Liu, H., Yuan, M., He, W.T., Beutler, N., Zhu, X., Tse, L.V., Martinez, D.R., Schafer, A., Anzanello, F., Yong, P., Peng, L., Dueker, K., Musharrafieh, R., Callaghan, S., Capozzola, T., Limbo, O., Parren, M., Garcia, E., Rawlings, S.A., Smith, D.M., Nemazee, D., Jardine, J.G., Safonova, Y., Briney, B., Rogers, T.F., Wilson, I.A., Baric, R.S., Gralinski, L.E., Burton, D.R., Andrabi, R.(2023) Immunity 56: 669-686.e7
- PubMed: 36889306
- DOI: https://doi.org/10.1016/j.immuni.2023.02.005
- Primary Citation of Related Structures:
8DGU, 8DGV, 8DGW, 8DGX - PubMed Abstract:
Pan-betacoronavirus neutralizing antibodies may hold the key to developing broadly protective vaccines against novel pandemic coronaviruses and to more effectively respond to SARS-CoV-2 variants. The emergence of Omicron and subvariants of SARS-CoV-2 illustrates the limitations of solely targeting the receptor-binding domain (RBD) of the spike (S) protein. Here, we isolated a large panel of broadly neutralizing antibodies (bnAbs) from SARS-CoV-2 recovered-vaccinated donors, which targets a conserved S2 region in the betacoronavirus spike fusion machinery. Select bnAbs showed broad in vivo protection against all three deadly betacoronaviruses, SARS-CoV-1, SARS-CoV-2, and MERS-CoV, which have spilled over into humans in the past two decades. Structural studies of these bnAbs delineated the molecular basis for their broad reactivity and revealed common antibody features targetable by broad vaccination strategies. These bnAbs provide new insights and opportunities for antibody-based interventions and for developing pan-betacoronavirus vaccines.
Organizational Affiliation:
Department of Immunology and Microbiology, The Scripps Research Institute, La Jolla, CA 92037, USA; IAVI Neutralizing Antibody Center, The Scripps Research Institute, La Jolla, CA 92037, USA; Consortium for HIV/AIDS Vaccine Development (CHAVD), The Scripps Research Institute, La Jolla, CA 92037, USA.