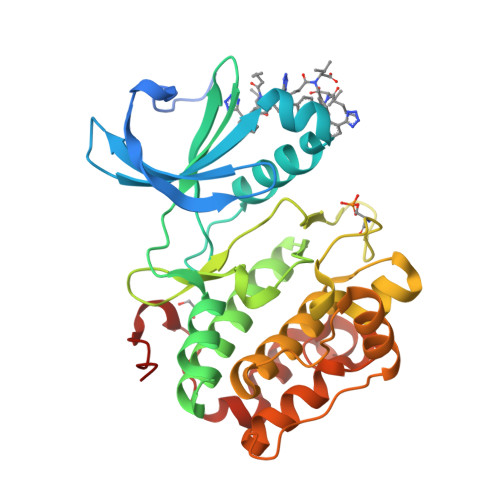
Modulation of the substrate specificity of the kinase PDK1 by distinct conformations of the full-length protein.
Sacerdoti, M., Gross, L.Z.F., Riley, A.M., Zehnder, K., Ghode, A., Klinke, S., Anand, G.S., Paris, K., Winkel, A., Herbrand, A.K., Godage, H.Y., Cozier, G.E., Suss, E., Schulze, J.O., Pastor-Flores, D., Bollini, M., Cappellari, M.V., Svergun, D., Grawert, M.A., Aramendia, P.F., Leroux, A.E., Potter, B.V.L., Camacho, C.J., Biondi, R.M.(2023) Sci Signal 16: eadd3184-eadd3184
- PubMed: 37311034
- DOI: https://doi.org/10.1126/scisignal.add3184
- Primary Citation of Related Structures:
8DQT - PubMed Abstract:
The activation of at least 23 different mammalian kinases requires the phosphorylation of their hydrophobic motifs by the kinase PDK1. A linker connects the phosphoinositide-binding PH domain to the catalytic domain, which contains a docking site for substrates called the PIF pocket. Here, we used a chemical biology approach to show that PDK1 existed in equilibrium between at least three distinct conformations with differing substrate specificities. The inositol polyphosphate derivative HYG8 bound to the PH domain and disrupted PDK1 dimerization by stabilizing a monomeric conformation in which the PH domain associated with the catalytic domain and the PIF pocket was accessible. In the absence of lipids, HYG8 potently inhibited the phosphorylation of Akt (also termed PKB) but did not affect the intrinsic activity of PDK1 or the phosphorylation of SGK, which requires docking to the PIF pocket. In contrast, the small-molecule valsartan bound to the PIF pocket and stabilized a second distinct monomeric conformation. Our study reveals dynamic conformations of full-length PDK1 in which the location of the linker and the PH domain relative to the catalytic domain determines the selective phosphorylation of PDK1 substrates. The study further suggests new approaches for the design of drugs to selectively modulate signaling downstream of PDK1.
Organizational Affiliation:
Instituto de Investigación en Biomedicina de Buenos Aires (IBioBA)-CONICET-Partner Institute of the Max Planck Society, Buenos Aires C1425FQD, Argentina.