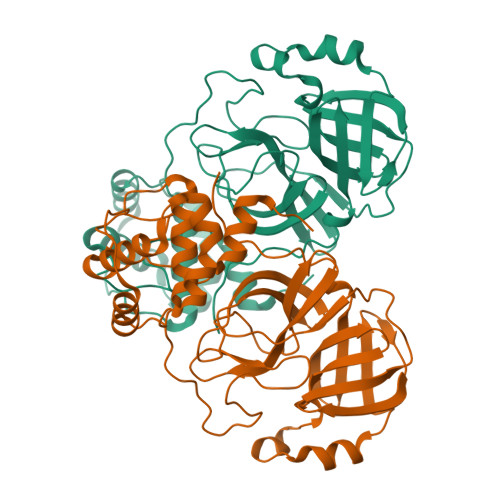
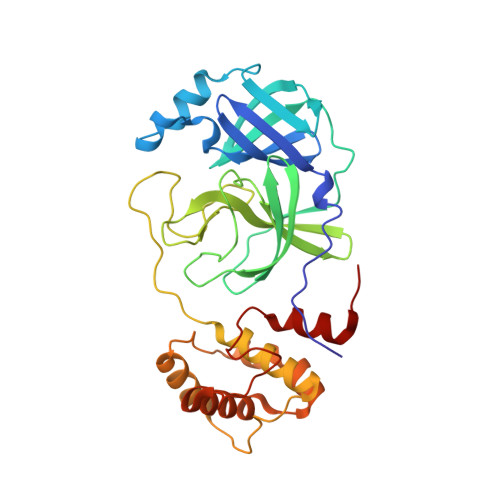
SARS-CoV-2 polyprotein substrate regulates the stepwise M pro cleavage reaction.
Narwal, M., Armache, J.P., Edwards, T.J., Murakami, K.S.(2023) J Biol Chem 299: 104697-104697
- PubMed: 37044215
- DOI: https://doi.org/10.1016/j.jbc.2023.104697
- Primary Citation of Related Structures:
8EIR, 8EKE - PubMed Abstract:
The processing of the Coronavirus polyproteins pp1a and pp1ab by the main protease M pro to produce mature proteins is a crucial event in virus replication and a promising target for antiviral drug development. M pro cleaves polyproteins in a defined order, but how M pro and/or the polyproteins determine the order of cleavage remains enigmatic due to a lack of structural information about polyprotein-bound M pro . Here, we present the cryo-EM structures of SARS-CoV-2 M pro in an apo form and in complex with the nsp7-10 region of the pp1a polyprotein. The complex structure shows that M pro interacts with only the recognition site residues between nsp9 and nsp10, without any association with the rest of the polyprotein. Comparison between the apo form and polyprotein-bound structures of M pro highlights the flexible nature of the active site region of M pro , which allows it to accommodate ten recognition sites found in the polyprotein. These observations suggest that the role of M pro in selecting a preferred cleavage site is limited and underscores the roles of the structure, conformation, and/or dynamics of the polyproteins in determining the sequence of polyprotein cleavage by M pro .
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Pennsylvania State University, University Park, Pennsylvania, USA.