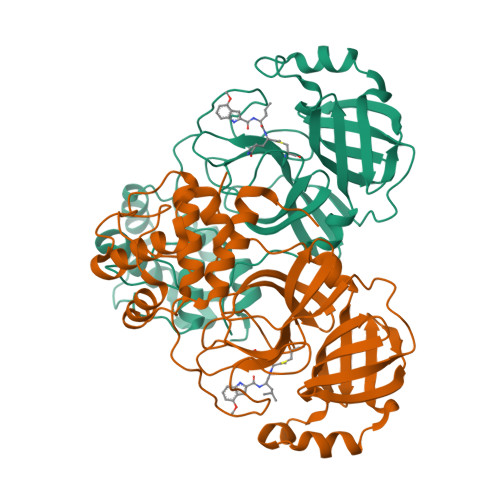
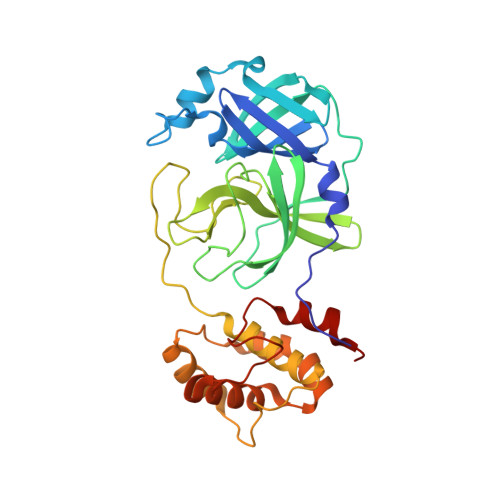
Alkyne as a Latent Warhead to Covalently Target SARS-CoV-2 Main Protease.
Ngo, C., Fried, W., Aliyari, S., Feng, J., Qin, C., Zhang, S., Yang, H., Shanaa, J., Feng, P., Cheng, G., Chen, X.S., Zhang, C.(2023) J Med Chem 66: 12237-12248
- PubMed: 37595260
- DOI: https://doi.org/10.1021/acs.jmedchem.3c00810
- Primary Citation of Related Structures:
8FY6, 8FY7 - PubMed Abstract:
There is an urgent need for improved therapy to better control the ongoing COVID-19 pandemic. The main protease M pro plays a pivotal role in SARS-CoV-2 replications, thereby representing an attractive target for antiviral development. We seek to identify novel electrophilic warheads for efficient, covalent inhibition of M pro . By comparing the efficacy of a panel of warheads installed on a common scaffold against M pro , we discovered that the terminal alkyne could covalently modify M pro as a latent warhead. Our biochemical and X-ray structural analyses revealed the irreversible formation of the vinyl-sulfide linkage between the alkyne and the catalytic cysteine of M pro . Clickable probes based on the alkyne inhibitors were developed to measure target engagement, drug residence time, and off-target effects. The best alkyne-containing inhibitors potently inhibited SARS-CoV-2 infection in cell infection models. Our findings highlight great potentials of alkyne as a latent warhead to target cystine proteases in viruses and beyond.
Organizational Affiliation:
Department of Chemistry and Loker Hydrocarbon Research Institute, University of Southern California, Los Angeles, California 90089, United States.