In Vitro Pharmacological Profile of KW-6356, a Novel Adenosine A 2A Receptor Antagonist/Inverse Agonist.
Ohno, Y., Suzuki, M., Asada, H., Kanda, T., Saki, M., Miyagi, H., Yasunaga, M., Suno, C., Iwata, S., Saito, J.I., Uchida, S.(2023) Mol Pharmacol 103: 311-324
- PubMed: 36894319
- DOI: https://doi.org/10.1124/molpharm.122.000633
- Primary Citation of Related Structures:
8GNE, 8GNG - PubMed Abstract:
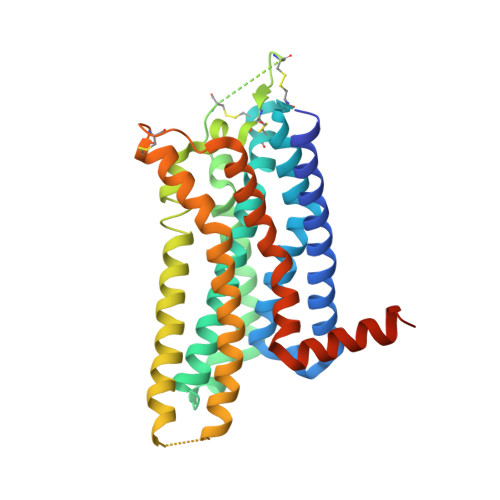
KW-6356 is a novel adenosine A 2A (A 2A ) receptor antagonist/inverse agonist, and its efficacy as monotherapy in Parkinson's disease (PD) patients has been reported. Istradefylline is a first-generation A 2A receptor antagonist approved for use as adjunct treatment to levodopa/decarboxylase inhibitor in adult PD patients experiencing "OFF" episodes. In this study, we investigated the in vitro pharmacological profile of KW-6356 as an A 2A receptor antagonist/inverse agonist and the mode of antagonism and compared them with istradefylline. In addition, we determined cocrystal structures of A 2A receptor in complex with KW-6356 and istradefylline to explore the structural basis of the antagonistic properties of KW-6356. Pharmacological studies have shown that KW-6356 is a potent and selective ligand for the A 2A receptor (the -log of inhibition constant = 9.93 ± 0.01 for human receptor) with a very low dissociation rate from the receptor (the dissociation kinetic rate constant = 0.016 ± 0.006 minute -1 for human receptor). In particular, in vitro functional studies indicated that KW-6356 exhibits insurmountable antagonism and inverse agonism, whereas istradefylline exhibits surmountable antagonism. Crystallography of KW-6356- and istradefylline-bound A 2A receptor have indicated that interactions with His250 6.52 and Trp246 6.48 are essential for the inverse agonism, whereas the interactions at both deep inside the orthosteric pocket and the pocket lid stabilizing the extracellular loop conformation may contribute to the insurmountable antagonism of KW-6356. These profiles may reflect important differences in vivo and help predict better clinical performance. SIGNIFICANCE STATEMENT: KW-6356 is a potent and selective adenosine A 2A receptor antagonist/inverse agonist and exhibits insurmountable antagonism, whereas istradefylline, a first-generation adenosine A 2A receptor antagonist, exhibits surmountable antagonism. Structural studies of adenosine A 2A receptor in complex with KW-6356 and istradefylline explain the characteristic differences in the pharmacological properties of KW-6356 and istradefylline.
Organizational Affiliation:
Biomedical Science Research Laboratories 1 (Y.O., S.U.) and Molecular Analysis Center (Mi.S., H.M., J.S.), Research Unit, R&D Division, Kyowa Kirin Co., Ltd., Shizuoka, Japan; Department of Cell Biology, Graduate School of Medicine, Kyoto University, Kyoto, Japan (H.A., S.I.); R&D Planning Department, R&D Division, Kyowa Kirin Co., Ltd., Tokyo, Japan (T.K.); Medical Affairs Department, Kyowa Kirin Co., Ltd., Tokyo, Japan (Ma.S.); CMC R&D Center, Production Division, Kyowa Kirin Co., Ltd., Shizuoka, Japan (M.Y.); and Department of Medical Chemistry, Kansai Medical University, Osaka, Japan (C.S.).