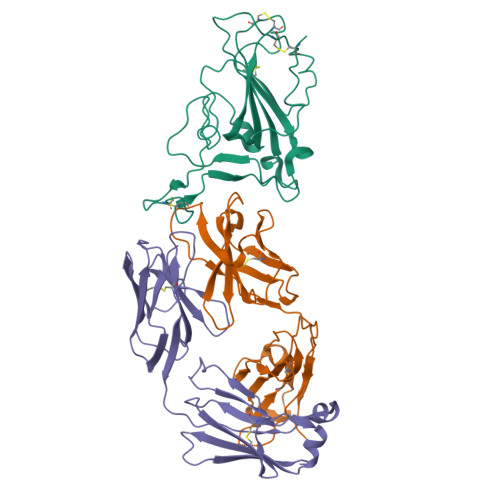
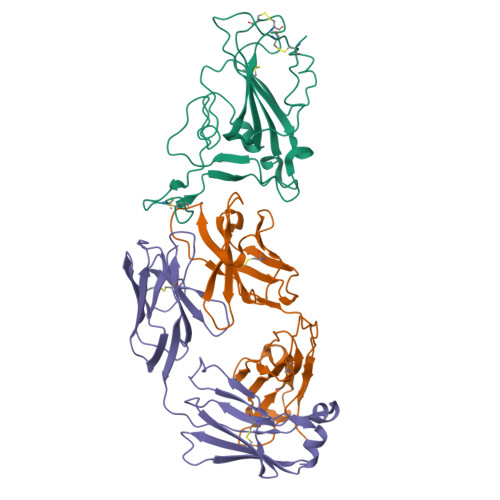
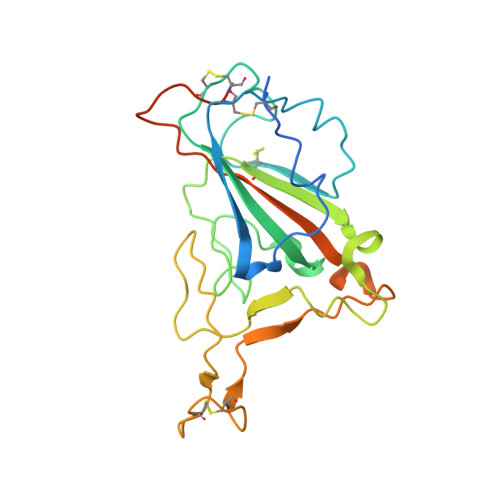
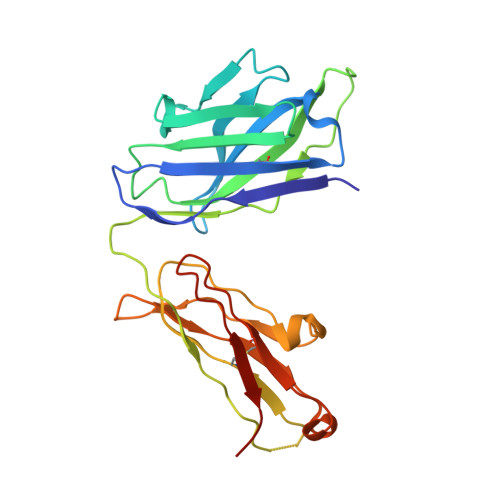
Structural basis of spike RBM-specific human antibodies counteracting broad SARS-CoV-2 variants.
Shitaoka, K., Higashiura, A., Kawano, Y., Yamamoto, A., Mizoguchi, Y., Hashiguchi, T., Nishimichi, N., Huang, S., Ito, A., Ohki, S., Kanda, M., Taniguchi, T., Yoshizato, R., Azuma, H., Kitajima, Y., Yokosaki, Y., Okada, S., Sakaguchi, T., Yasuda, T.(2023) Commun Biol 6: 395-395
- PubMed: 37041231
- DOI: https://doi.org/10.1038/s42003-023-04782-6
- Primary Citation of Related Structures:
7WN2, 7WNB, 7YOW, 8I5H, 8I5I - PubMed Abstract:
The decrease of antibody efficacy to mutated SARS-CoV-2 spike RBD explains the breakthrough infections and reinfections by Omicron variants. Here, we analyzed broadly neutralizing antibodies isolated from long-term hospitalized convalescent patients of early SARS-CoV-2 strains. One of the antibodies named NCV2SG48 is highly potent to broad SARS-CoV-2 variants including Omicron BA.1, BA.2, and BA.4/5. To reveal the mode of action, we determined the sequence and crystal structure of the Fab fragment of NCV2SG48 in a complex with spike RBD from the original, Delta, and Omicron BA.1. NCV2SG48 is from a minor V H but the multiple somatic hypermutations contribute to a markedly extended binding interface and hydrogen bonds to interact with conserved residues at the core receptor-binding motif of RBD, which efficiently neutralizes a broad spectrum of variants. Thus, eliciting the RBD-specific B cells to the longitudinal germinal center reaction confers potent immunity to broad SARS-CoV-2 variants emerging one after another.
Organizational Affiliation:
Department of Immunology, Graduate School of Biomedical and Health Sciences, Hiroshima University, Hiroshima, Japan.