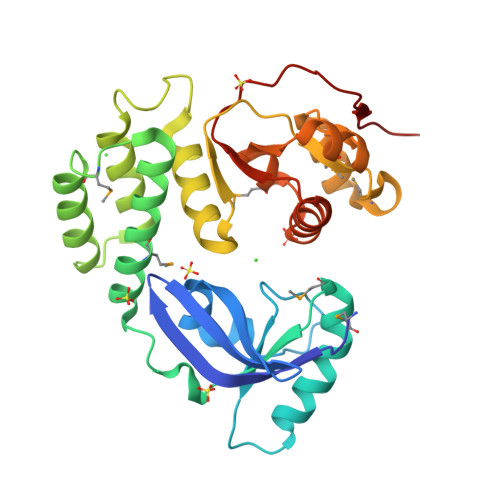
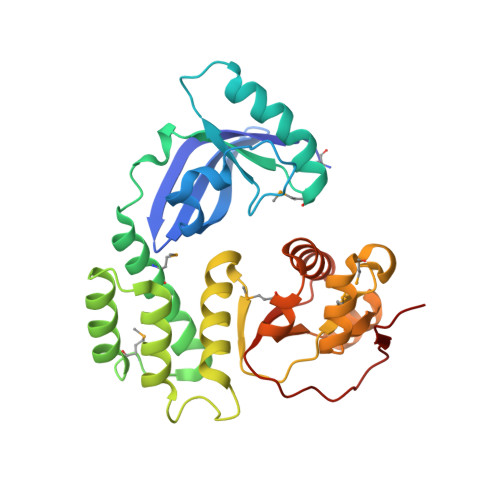
The CoV-Y domain of SARS-CoV-2 Nsp3 interacts with BRAP to stimulate NF-kappa B signaling and induce host inflammatory responses.
Wang, K., Ni, X., Deng, X., Nan, J., Ma-Lauer, Y., von Brunn, A., Zeng, R., Lei, J.(2024) Int J Biol Macromol 280: 136123-136123
- PubMed: 39343285
- DOI: https://doi.org/10.1016/j.ijbiomac.2024.136123
- Primary Citation of Related Structures:
8ILC - PubMed Abstract:
Non-structural protein 3 (Nsp3) is the largest protein encoded by the coronavirus (CoV) genome. It consists of multiple domains that perform critical functions during the viral life cycle. CoV-Y is the most C-terminal domain of Nsp3, and it exhibits evolutionary conservation across diverse CoVs; however, the exact biological function of CoV-Y remains unclear. Here, we determined the crystal structure of CoV-Y of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Nsp3 using the single-wavelength anomalous diffraction method. We revealed the interaction between CoV-Y and the host BRCA1-associated protein (BRAP) using immunoprecipitation-mass spectrometry experiments. This interaction was subsequently confirmed in cellular assays, and the precise binding-regions between these two proteins were clarified. We found that this interaction is conserved in SARS-CoV and Middle East respiratory syndrome coronavirus. Next, we demonstrated that CoV-Y enhances IκBα and IκBβ phosphorylation and promotes the nuclear translocation of the downstream NF-κB members p50 and p65 through binding to BRAP. The CoV-Y-BRAP interaction can upregulate the transcript levels of the host inflammatory cytokines. Overall, our findings illustrate the biological function of CoV-Y for the first time and provide novel insights into coronavirus regulation of host inflammatory responses, as well as a possible target for antiviral drug development.
Organizational Affiliation:
National Clinical Research Center for Geriatrics, and State Key Laboratory of Biotherapy, West China Hospital, Sichuan University, Chengdu, Sichuan, China.