SignatureFinder enables sequence mining to identify cobalamin-dependent photoreceptor proteins
Yu, Y., Jeffreys, L.N., Poddar, H., Hill, A., Johannissen, L., Dai, F., Sakuma, M., Leys, D., Heyes, D.J., Zhang, S., Scrutton, N.S.To be published.
Experimental Data Snapshot
Starting Model: in silico
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Putative cobalamin binding protein | 327 | Chloracidobacterium thermophilum | Mutation(s): 0 | 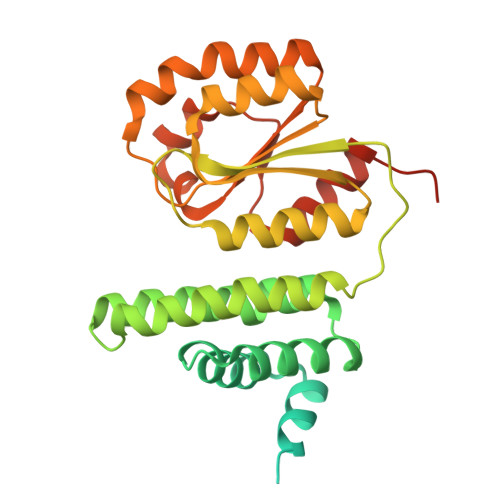 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| B12 (Subject of Investigation/LOI) Query on B12 | C [auth A], E [auth B] | COBALAMIN C62 H89 Co N13 O14 P LKVIQTCSMMVGFU-DWSMJLPVSA-N |  | ||
| 5AD (Subject of Investigation/LOI) Query on 5AD | D [auth A], F [auth B] | 5'-DEOXYADENOSINE C10 H13 N5 O3 XGYIMTFOTBMPFP-KQYNXXCUSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 125.218 | α = 90 |
| b = 125.218 | β = 90 |
| c = 73.088 | γ = 120 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| DIALS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Engineering and Physical Sciences Research Council | United Kingdom | EP/S030336/1 |