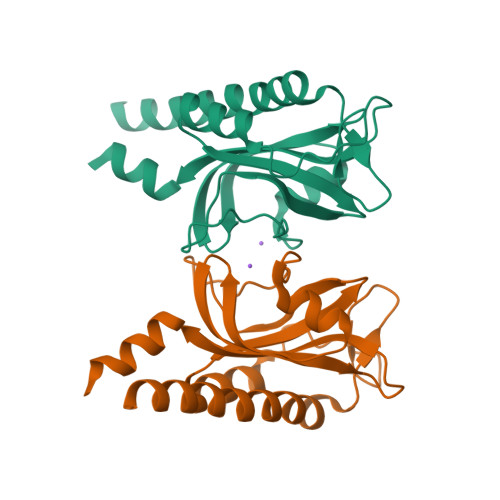
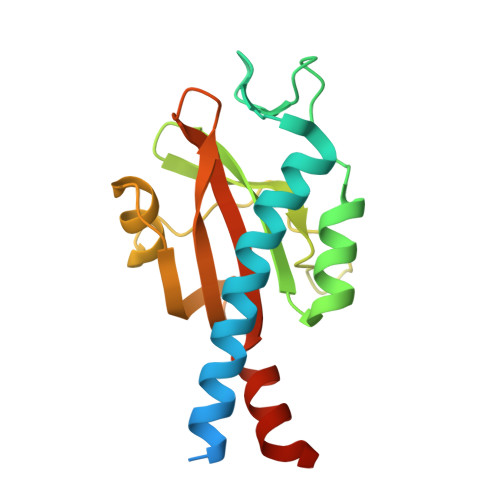
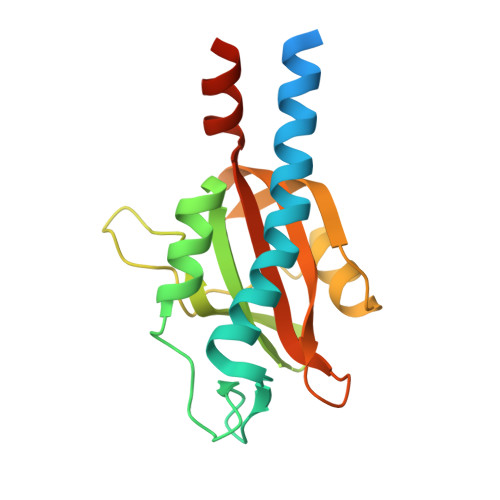
Bacterial sensor evolved by decreasing complexity.
Monteagudo-Cascales, E., Gavira, J.A., Xing, J., Velando, F., Matilla, M.A., Zhulin, I.B., Krell, T.(2025) Proc Natl Acad Sci U S A 122: e2409881122-e2409881122
- PubMed: 39879239
- DOI: https://doi.org/10.1073/pnas.2409881122
- Primary Citation of Related Structures:
8PY0, 8PY1 - PubMed Abstract:
Bacterial receptors feed into multiple signal transduction pathways that regulate a variety of cellular processes including gene expression, second messenger levels, and motility. Receptors are typically activated by signal binding to ligand-binding domains (LBDs). Cache domains are omnipresent LBDs found in bacteria, archaea, and eukaryotes, including humans. They form the predominant family of extracytosolic bacterial LBDs and were identified in all major receptor types. Cache domains are composed of either a single (sCache) or a double (dCache) structural module. The functional relevance of bimodular LBDs remains poorly understood. Here, we identify the PacF chemoreceptor in the phytopathogen Pectobacterium atrosepticum that recognizes formate at the membrane-distal module of its dCache domain, triggering chemoattraction. We further demonstrate that a family of formate-specific sCache domains has evolved from a dCache domain, exemplified by PacF, by losing the membrane-proximal module. By solving high-resolution structures of two family members in complex with formate, we show that the molecular basis for formate binding at sCache and dCache domains is highly similar, despite their low sequence identity. The apparent loss of the membrane-proximal module may be related to the observation that dCache domains bind ligands typically at the membrane-distal module, whereas studies have failed to find ligands bound in the membrane-proximal module. This work advances our understanding of signal sensing in bacterial receptors and suggests that evolution by reducing complexity may be a route for shaping diversity.
Organizational Affiliation:
Department of Biotechnology and Environmental Protection, Estación Experimental del Zaidín, Consejo Superior de Investigaciones Científicas, Granada 18008, Spain.