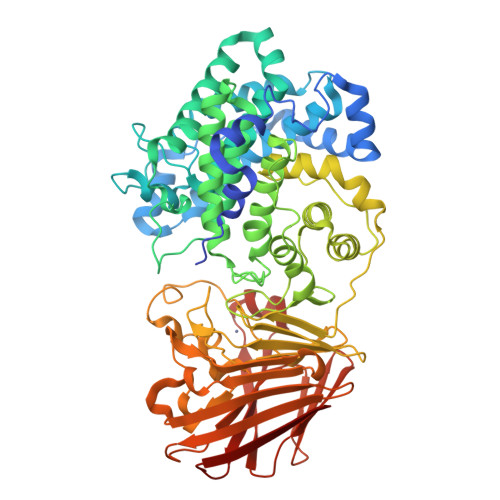
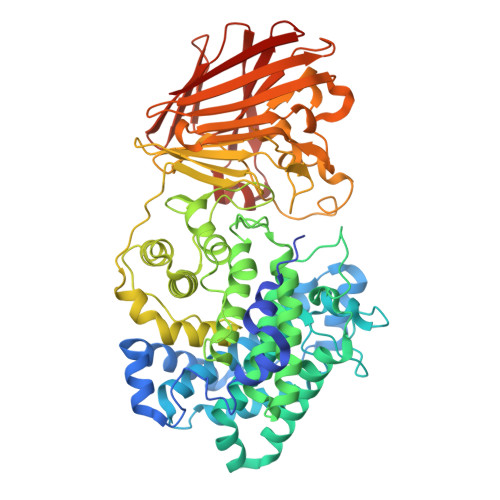
Bacterial polysaccharide lyase family 33: Specificity from an evolutionarily conserved binding tunnel.
Loiodice, M., Drula, E., McIver, Z., Antonyuk, S., Basle, A., Lima, M., Yates, E.A., Byrne, D.P., Coughlan, J., Leech, A., Mesdaghi, S., Rigden, D.J., Drouillard, S., Helbert, W., Henrissat, B., Terrapon, N., Wright, G.S.A., Couturier, M., Cartmell, A.(2025) Proc Natl Acad Sci U S A 122: e2421623122-e2421623122
- PubMed: 39932998
- DOI: https://doi.org/10.1073/pnas.2421623122
- Primary Citation of Related Structures:
8R6Z, 8R70, 8R71, 8R72, 8R73, 8R75 - PubMed Abstract:
Acidic glycans are essential for the biology of multicellular eukaryotes. To utilize them, microbial life including symbionts and pathogens has evolved polysaccharide lyases (PL) that cleave their 1,4 glycosidic linkages via a β-elimination mechanism. PL family 33 (PL33) enzymes have the unusual ability to target a diverse range of glycosaminoglycans (GAGs), as well as the bacterial polymer, gellan gum. In order to gain more detailed insight into PL33 activities we recombinantly expressed 10 PL33 members derived from all major environments and further elucidated the detailed biochemical and biophysical properties of five, showing that their substrate specificity is conferred by variations in tunnel length and topography. The key amino acids involved in catalysis and substrate interactions were identified, and employing a combination of complementary biochemical, structural, and modeling approaches, we show that the tunnel topography is induced by substrate binding to the glycan. Structural and bioinformatic analyses revealed that these features are conserved across several lyase families as well as in mammalian GAG epimerases.
Organizational Affiliation:
Université Grenoble Alpes, CNRS, Centre de Recherche sur les Macromolécules Végétales, Grenoble 38000, France.