Phage parasites targeting phage homologous recombinases provide antiviral immunity.
Debiasi-Anders, G., Qiao, C., Salim, A., Li, N., Mir-Sanchis, I.(2025) Nat Commun 16: 1889-1889
- PubMed: 39987160
- DOI: https://doi.org/10.1038/s41467-025-57156-3
- Primary Citation of Related Structures:
8PQ8, 8Q86, 8QE9, 8RC5 - PubMed Abstract:
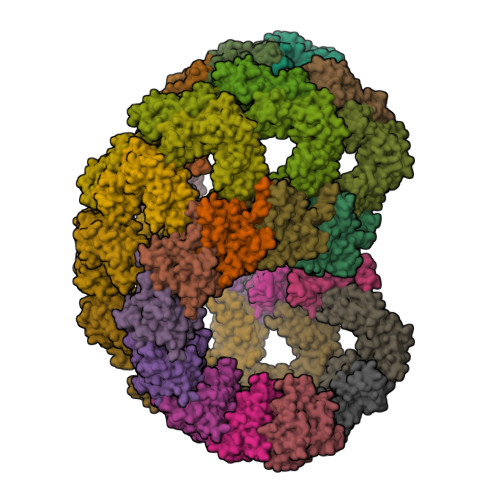
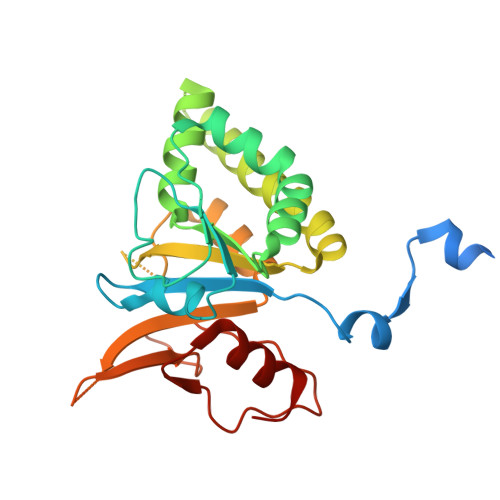
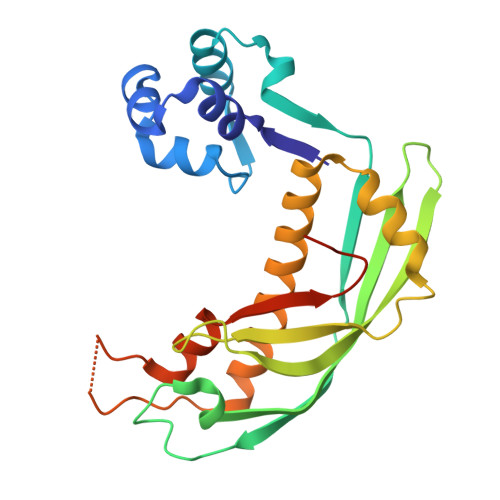
Bacteria often carry multiple genes encoding anti-phage defense systems, clustered in defense islands and phage satellites. Various unrelated anti-phage defense systems target phage-encoded homologous recombinases (HRs) through unclear mechanisms. Here, we show that the phage satellite SaPI2, which does not encode orthodox anti-phage defense systems, provides antiviral immunity mediated by Stl2, the SaPI2-encoded transcriptional repressor. Stl2 targets and inhibits phage-encoded HRs, including Sak and Sak4, two HRs from the Rad52-like and Rad51-like superfamilies. Remarkably, apo Stl2 forms a collar of dimers oligomerizing as closed rings and as filaments, mimicking the quaternary structure of its targets. Stl2 decorates both Sak rings and Sak4 filaments. The oligomerization of Stl2 as a collar of dimers is necessary for its inhibitory activity both in vitro and in vivo. Our results shed light on the mechanisms underlying antiviral immunity against phages carrying divergent HRs.
Organizational Affiliation:
Department of Medical Biochemistry and Biophysics, Umeå University, Umeå, Sweden.