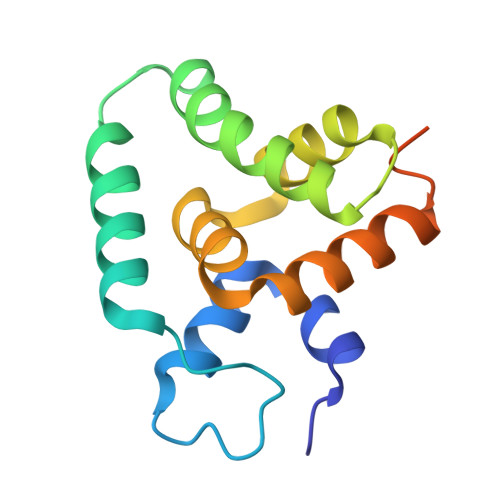
Deciphering the RNA-binding protein network during endosomal mRNA transport.
Devan, S.K., Shanmugasundaram, S., Muntjes, K., Postma, J., Smits, S.H.J., Altegoer, F., Feldbrugge, M.(2024) Proc Natl Acad Sci U S A 121: e2404091121-e2404091121
- PubMed: 39499630
- DOI: https://doi.org/10.1073/pnas.2404091121
- Primary Citation of Related Structures:
8S6N, 8S6O, 8S6U - PubMed Abstract:
Microtubule-dependent endosomal transport is crucial for polar growth, ensuring the precise distribution of cellular cargos such as proteins and mRNAs. However, the molecular mechanism linking mRNAs to the endosomal surface remains poorly understood. Here, we present a structural analysis of the key RNA-binding protein Rrm4 from Ustilago maydis . Our findings reveal a different type of MademoiseLLE domain (MLLE) featuring a seven-helical bundle that provides a distinct binding interface. A comparative analysis with the canonical MademoiseLLE domain of the poly(A)-binding protein Pab1 disclosed unique characteristics of both domains. Deciphering the MLLE binding code enabled prediction and verification of previously unknown Rrm4 interactors containing short linear motifs. Importantly, we demonstrated that the human MLLE domains, such as those of PABPC1 and UBR5, employed a similar principle to distinguish among interaction partners. Thus, our study provides detailed mechanistic insights into how structural variations in the widely distributed MLLE domain facilitate mRNA attachment during endosomal transport.
Organizational Affiliation:
Department of Biology, Institute of Microbiology, Cluster of Excellence on Plant Sciences, Heinrich Heine University Düsseldorf, Düsseldorf 40204, Germany.