A unique binding mode of P1' Leu-containing target sequences for Streptococcus pyogenes sortase A results in alternative cleavage.
Vogel, B.A., Blount, J.M., Kodama, H.M., Goodwin-Rice, N.J., Andaluz, D.J., Jackson, S.N., Antos, J.M., Amacher, J.F.(2024) RSC Chem Biol 5: 30-40
- PubMed: 38179192
- DOI: https://doi.org/10.1039/d3cb00129f
- Primary Citation of Related Structures:
8T8G - PubMed Abstract:
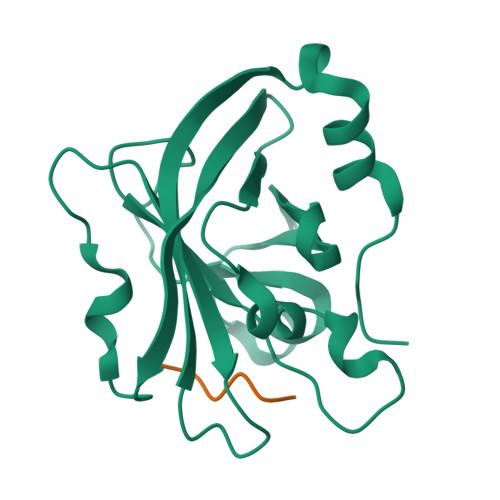
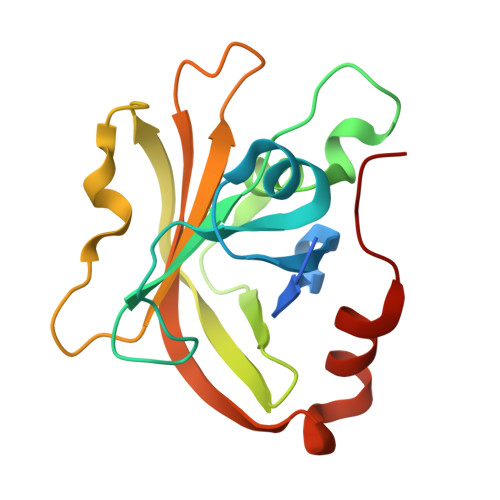
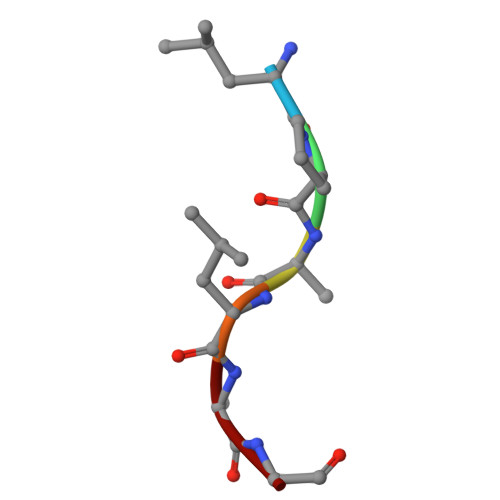
Sortase enzymes are cysteine transpeptidases that attach environmental sensors, toxins, and other proteins to the cell surface in Gram-positive bacteria. The recognition motif for many sortases is the cell wall sorting signal (CWSS), LPXTG, where X = any amino acid. Recent work from ourselves and others has described recognition of additional amino acids at a number of positions in the CWSS, specifically at the Thr (or P1) and Gly (or P1') positions. In addition, although standard cleavage occurs between these two residues (P1/P1'), we previously observed that the SrtA enzyme from Streptococcus pneumoniae will cleave after the P1' position when its identity is a Leu or Phe. The stereochemical basis of this alternative cleavage is not known, although homologs, e.g. , SrtA from Listeria monocytogenes or Staphylococcus aureus do not show alternative cleavage to a significant extent. Here, we use protein biochemistry, structural biology, and computational biochemistry to predict an alternative binding mode that facilitates alternative cleavage. We use Streptococcus pyogenes SrtA (spySrtA) as our model enzyme, first confirming that it shows similar standard/alternative cleavage ratios for LPAT L , LPAT F , and LPAT Y sequences. Molecular dynamics simulations suggest that when P1' is Leu, this amino acid binds in the canonical S1 pocket, pushing the P1 Thr towards solvent. The P4 Leu (L̲PATL) binds as it does in standard binding, resulting in a puckered binding conformation. We use P1 Glu-containing peptides to support our hypotheses, and present the complex structure of spySrtA-LPALA to confirm favorable accommodation of Leu in the S1 pocket. Overall, we structurally characterize an alternative binding mode for spySrtA and specific target sequences, expanding the potential protein engineering possibilities in sortase-mediated ligation applications.
Organizational Affiliation:
Department of Chemistry, Western Washington University, 516 High St - MS9150 Bellingham WA 98225 USA antosj@wwu.edu amachej@wwu.edu +1-360-650-2826 +1-360-650-2271 +1-360-650-4397.